Introduction
Frontotemporal dementia (FTD) is a degenerative disease that primarily affects the frontal and anterior temporal lobes. It is the third most common form of primary dementia, following Alzheimer’s disease and Lewy body dementia. While the exact cause of FTD remains unclear, genetic factors play a significant role, accounting for approximately 40% of cases. The disease is largely driven by these genetic factors and the degeneration of neurons in these regions. Patients typically experience a decline in various functions, particularly language and social behavior. FTD is a type of early-onset dementia that usually manifests between the ages of 45 and 64, with an average onset around 57.1 The incidence is roughly equal between men and women. Unlike Alzheimer’s disease, which predominantly affects memory, FTD rarely involves memory impairment. Instead, early signs include behavioral abnormalities, deficits in executive function, and language difficulties, making it challenging for patients to recognize the condition earlier on.2
Currently, FTD cannot be completely cured, and drug development is still in progress. Only a few drugs have been tested in live mouse models, typically by observing whether early injection of the drug in mice with frontotemporal degeneration can prevent the occurrence of motor function abnormalities.3 Common drugs used to treat FTD include rapamycin, spermidine, carbamazepine, tamoxifen, and other autophagy activators. These drugs help reduce the overexpression of TDP-43 protein.4 Rapamycin, an mTOR inhibitor, is a central regulator of cell growth, controlling various cellular processes. In tumor cells, this regulatory mechanism can become dysregulated, leading to uncontrolled cell growth. Therefore, rapamycin is used to inhibit mTOR, helping to restore normal cellular function and prevent tumor progression.5 In addition to rapamycin, drugs such as spermidine, carbamazepine, and tamoxifen can provide some control over FTD. Some individuals may also take antidepressants or antipsychotic medications to alleviate behavioral and emotional issues associated with FTD. However, these come with significant side effects, including neurological damage, drowsiness, dizziness. Currently, common therapies for FTD also include supportive therapies, speech therapy, and cognitive-behavioral therapy. These methods focus on improving patients’ quality of life by controlling symptoms and enhancing communication and cognitive abilities.6
As noted above, there is currently no single drug or treatment that can completely cure FTD. However, the drug discovery process integrates computational techniques, experimental validation, translational models, and clinical trials to uncover potential therapeutic candidates. Despite considerable progress in biotechnology and an enhanced comprehension of biological systems, drug discovery remains an expensive, lengthy, and inefficient endeavor, with a high failure rate in developing new treatments.7 Only about 10–20% of candidate drugs successfully progress from the start of clinical trials to market approval, a figure that has remained largely unchanged for decades.8 Thus, there is a pressing need for a more efficient and systematic approach to drug design. Drug-target interaction (DTI) is a crucial aspect of the drug development process. When a drug binds to its target, such as a protein or gene, it alters the target’s biological activity, helping to restore normal function. Predicting drug-target interactions is vital in drug discovery, as it can improve efficiency and reduce costs.9
DTI prediction often involves four main types of targets: proteins, diseases, genes, and side effects.10 Discovering new targets for existing or discontinued drugs—a process known as drug repurposing—is another important aspect of drug discovery. With advancements in pharmacology, the ‘multi-target, multi-drug’ model has gained widespread acceptance, replacing the traditional ‘one target, one drug’ approach. One key insight is that drugs often target multiple proteins rather than focusing on just one. Therefore, multi-molecular combination drugs are a current trend in drug development. These drugs can work synergistically to enhance the effectiveness of each drug in the combination and help reduce drug resistance, toxicity, and adverse reactions.11
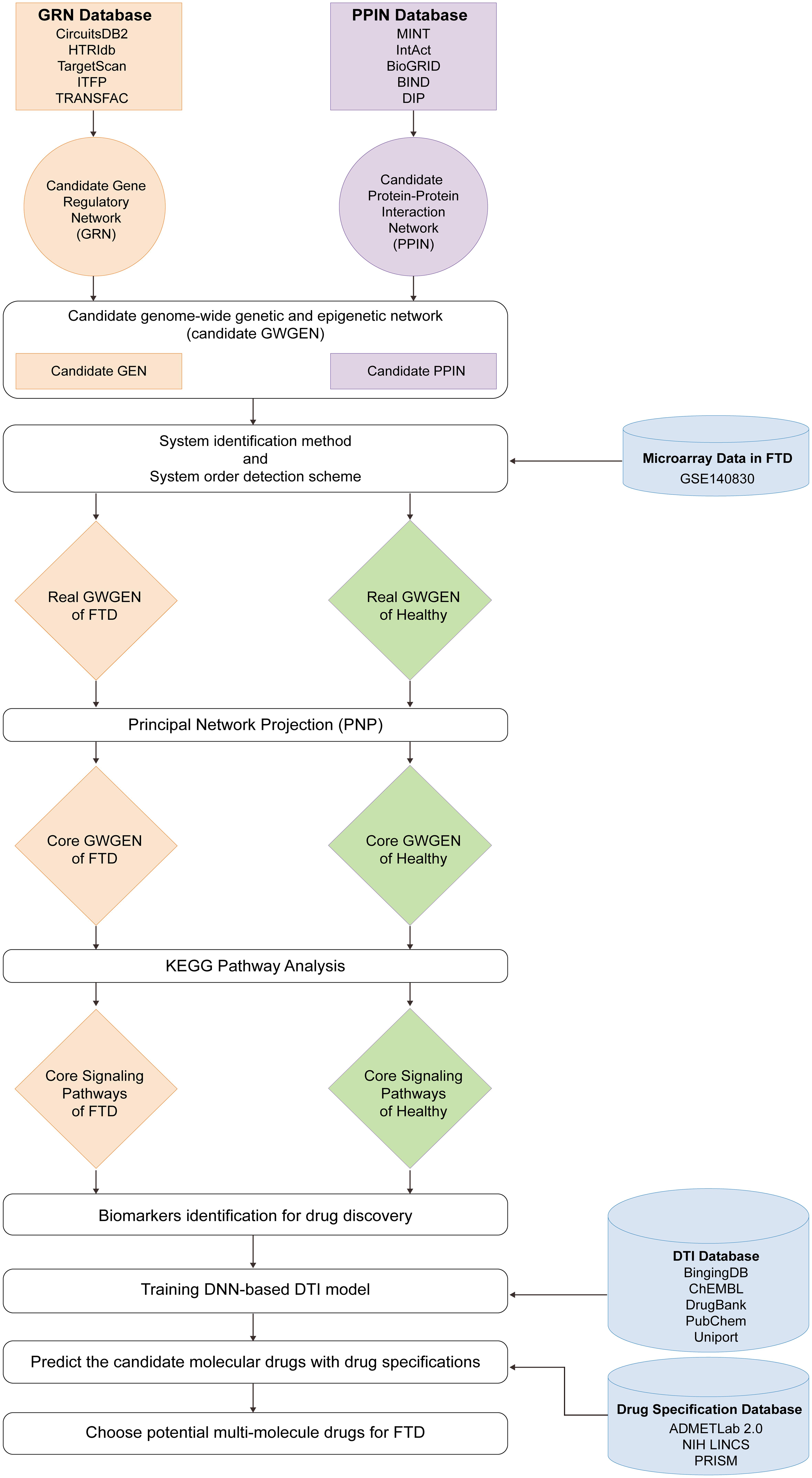
In this study, we developed a workflow, illustrated in Figure 1. Based on the previous discussion, the ‘multi-target, multi-drug’ model is the prevailing approach. Our primary goal is to optimize multi-target treatment strategies by designing drugs that can simultaneously target multiple biological pathways, maximizing therapeutic efficacy. For example, FTD involves multiple biological signaling pathways and pathological mechanisms, so multi-target drugs are better suited to inhibit various aspects of the disease. Additionally, our research aimed to enhance efficacy while minimizing drug resistance. To address drug resistance, we are investigating how simultaneous interference with multiple targets can reduce the likelihood of pathogens or tumor cells developing resistance. The diversity of targets in multi-molecular drugs makes it more difficult for pathogens or cancer cells to evade treatment, thereby improving overall efficacy.
(I) Big data mining from gene regulatory network (GRN) and protein-protein interaction network (PPIN) databases to construct candidate genome-wide genetic and epigenetic networks (GWGENs); (II) Microarray data of FTD and healthy control are employed to construct real GWGENs of FTD and healthy control using a system identification method; (III) Principal network projection (PNP) method is used to extract core GWGENs from real GWGENs for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation to identify the core signaling pathways of FTD and healthy control and to identify significant biomarkers; (IV) A deep neural network (DNN)-based drug-target interaction (DTI) model, trained with DTI databases, is used to predict candidate molecular drugs, which are screened based on drug design specifications to obtain potential multi-molecule drugs for FTD.
Materials and methods
To construct the core genome-wide genetic and epigenetic networks (GWGENs), we downloaded the microarray data from the GSE140830 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140830 ) available through the National Center for Biotechnology Information. This dataset includes data from FTD patients and healthy control. For data preprocessing, we first divided the original dataset into five categories: gene, transcription factor (TF), receptor, lncRNA, and miRNA, and then proceeded to rank them.
Research ethics
Ethical approval is not applicable due to the use of the publicly available dataset GSE140830 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140830 ).
Using this dataset, we constructed the core GWGENs step by step, as shown in Figure 1.
(I) Construction of Candidate GWGENs: The first step involves creating candidate GWGENs, which include the candidate Protein-Protein Interaction Network (PPIN) and the candidate Gene Regulatory Network (GRN) identified through tree-based mining methods.
(II) Identification of Real GWGENs: To remove false positive data, we established all possible regression systems for each node and used the least squares method for system identification. This method applies a system order detection approach to identify the real GWGENs for FTD and healthy control using whole genome microarray data.
(III) Extraction of Core GWGENs: We employed the Principal Network Projection (PNP) method, which uses singular value decomposition to select the top 6,000 nodes with the highest projection values in the GWGENs. These 6,000 nodes, which exhibit the strongest projections on significant singular vector structures accounting for 85% of the real GWGENs, are considered the core GWGENs for further analysis.
(IV) Designing a multi-molecule drug for the treatment of FTD: After identifying the core GWGENs, we annotated them using KEGG pathways to identify core signaling pathways in FTD and healthy control. Based on various research records, we selected significant biomarkers related to critical pathogenic mechanisms as drug targets, i.e., TAU, GSK-3β, STAT3, ATG5, WDR41, and RIPK1. Using the deep neural network (DNN)-based DTI model trained on DTI databases, we predicted and screened potential molecular drugs by design specifications to combine them into a multi-molecule drug for treating FTD through targeting these significant biomarkers.
(I) Constructing the candidate GWGEN of FTD and healthy control through big data mining
In this study, the whole-genome microarray dataset with accession number GSE140830 was downloaded from the Gene Expression Omnibus at the National Center for Biotechnology Information. This dataset includes data from 234 blood samples of FTD patients and 248 blood samples of healthy control. Each sample contains expression levels of proteins, receptors, transcription factors, miRNAs, and lncRNAs. These sample data were preprocessed and mined using big data techniques to construct the candidate GWGEN. The candidate GWGEN involves logical and Boolean functions: an interaction between two nodes is recorded as 1, and the absence of interaction is recorded as 0. The candidate GWGEN is divided into the candidate PPIN and the candidate GRN. To construct the candidate PPIN, the following databases were used: MINT, IntAct, BioGRID, BIND, and DIP. To construct the candidate GRN, the following databases were used: CircuitsDB2, HTRIdb, TargetScan, ITFP, and TRANSFAC.
After completing the data preprocessing through the aforementioned databases, we constructed interactive and regulatory models for protein interactions and genetic regulations in the candidate GWGEN. These models included interactions among proteins and regulations among genes, transcription factors, miRNAs, and lncRNAs. The protein interaction and gene regulation models also accounted for random noise caused by model residuals and baseline levels.
For the protein-protein interaction (PPI) model, we constructed the following PPI equations:
In the protein-protein interaction model, Pi[n] and Pw[n] represent the expression levels of the i-th and w-th proteins in the n-th sample, respectively. The parameter σiw denotes the interaction strength between the i-th and w-th proteins. τi,PPIN represents the baseline expression level of the i-th protein due to unknown interactions caused by histone modifications, such as phosphorylation and acetylation. φi,PPIN[n] signifies the random measurement noise in the expression of the i-th protein in the n-th sample. Wi indicates the total number of interactions with the i-th protein. The letter I stands for the total number of proteins, and N represents the total number of samples.
For the GRN model, we constructed the following genetic regulatory equations:
In the GRN model, the terms gj[n], tx[n], ly[n], and mz[n] represent the expression levels of the j-th gene, x-th transcription factor, y-th lncRNA, and z-th miRNA in the n-th sample, respectively. The parameter εjx denotes the regulatory strength from the x-th transcription factor on the j-th gene. The parameter θjy represents the regulatory strength from the y-th lncRNA on the j-th gene. The parameter µjz denotes the regulatory strength from the z-th miRNA on the j-th gene, with µjz being a positive value due to the negative regulatory role of miRNAs on gene expression. The term τj signifies the baseline expression level of the j-th gene due to unknown regulations caused by histone modifications such as phosphorylation and acetylation. The term φj[n] represents the random noise in the measurement of expression of the j-th gene in the n-th sample. The terms Xj, Yj and Zj represent the total number of transcription factors, lncRNAs, and miRNAs regulating the j-th gene, respectively. The letter J stands for the total number of genes, and N represents the total number of samples.
For the lncRNA regulatory model, we constructed the following regulatory equations:
In the lncRNA regulatory model, the terms lk[n], tx[n], ly[n], and represent the expression levels of the k-th lncRNA, x-th transcription factor, y-th lncRNA, and z-th miRNA in the n-th sample, respectively. The parameter αkx denotes the regulatory strength from the x-th transcription factor on the k-th lncRNA. The parameter βky represents the regulatory strength from the y-th lncRNA on the k-th lncRNA. The parameter γkz denotes the regulatory strength from the z-th miRNA on the k-th lncRNA, with γkz being a positive value due to the negative regulatory role of miRNAs on lncRNA expression. The term τk signifies the baseline expression level of the k-th lncRNA due to unknown regulations caused by histone modifications such as phosphorylation and acetylation. The term φk[n] represents the random noise in the measurement of expression of the k-th lncRNA in the n-th sample. The terms Xk, Yk and Zk represent the total number of transcription factors, lncRNAs, and miRNAs regulating the k-th lncRNA, respectively. The letter K stands for the total number of lncRNAs, and N represents the total number of samples.
For the miRNA regulatory model, we constructed the following regulating equations:
In the miRNA regulatory model, the terms mh[n], tx[n], ly[n], and mz[n] represent the expression levels of the h-th miRNA, x-th transcription factor, y-th lncRNA, and z-th miRNA in the n-th sample, respectively. The parameter ηhx denotes the regulatory strength from the x-th transcription factor on the h-th miRNA. The parameter πhy represents the regulatory strength from the y-th lncRNA on the h-th miRNA. The parameter ξhz denotes the regulatory strength between the z-th miRNA and the h-th miRNA, with ξhz being a positive value due to the negative regulatory role of miRNAs on miRNA expression. The term τh signifies the baseline expression level of the h-th miRNA due to unknown regulations caused by histone modifications such as phosphorylation and acetylation. The term φh[n] represents the random noise in the measurement of expression of the h-th miRNA in the n-th sample. The terms Xh, Yh and Zh represent the total number of transcription factors, lncRNAs, and miRNAs regulating the h-th miRNA, respectively. The letter H stands for the total number of miRNAs, and N represents the total number of samples.
(II) Constructing the real GWGEN for FTD and healthy control using system identification and system order detection methods
In the previous section, we established four models for the candidate GWGEN, including proteins, genes, lncRNAs, and miRNAs. However, the candidate GWGEN only records whether there is an interaction or regulation between two nodes, while the actual expression levels vary from person to person. Furthermore, false positives may arise due to data mining from large databases. To eliminate false positives in the candidate GWGEN, we constructed the real GWGEN for FTD and the healthy controlby applying system identification and system order detection methods.
To determine the interaction and regulation parameters for the protein interaction and genetic regulatory models, we rewrote Equations (1) to (4) in linear regression form, as shown in Equations (5) to (8).
In these linear regression equations, the regression vectors ωi[n], ωj[n], ωk[n], and ωh[n] represent the expression levels of the i-th protein, j-th gene, k-th lncRNA, and h-th miRNA in the n-th sample, respectively. The parameter vector δi,PPIN denotes the interaction abilities of the i-th protein. The parameter vectors δj, δk, δh denote the regulatory abilities and basal levels of the j-th gene, k-th lncRNA, and h-th miRNA, respectively. The terms φi,PPIN[n], φj[n], φk[n], and φh [n] represent the random noise in the expression of the i-th protein, j-th gene, k-th lncRNA, and h-th miRNA in the n-th sample.
Equations (5) to (8) can be expanded by considering all sample data and rewritten as follows.
Equations (9) to (12) can be simplified as follows.
Equations (5) to (8) can be further expanded by considering all samples and rewritten as Equations (9) to (12). To avoid overfitting issues in the system identification methods for the random models, the number of elements in the parameters of the PPIN and GRN in Equations (13) to (16) (i.e., Δi, Δj, Δk, and Δh cannot exceed half of the dataset’s samples (N/2). Therefore, we determine the values of the parameter vectors Δi, Δj, Δk, and Δh by solving constrained linear least squares parameter estimation methods to ensure the negative regulatory role of miRNA in Equations (17) to (20).
Through Equations (17) to (20), we can obtain the optimal solutions for the parameter vectors
After addressing the constrained least square parameter estimation problems from the respective genome-wide microarray data, we obtained the interaction strengths among proteins in the candidate GWGEN for FTD and healthy control, as well as the regulatory abilities for genes, lncRNA, and miRNA. However, due to potential false positive errors in the data from various databases caused by different experimental conditions, we used the Akaike Information Criterion (AIC) to eliminate false positives and perform system order detection for each protein, gene, lncRNA, and miRNA. Therefore, we deleted the false positives from their system orders in the candidate GWGEN to obtain the real GWGEN, as shown in Figure S1.
Based on the system order detection model, we provided four different AIC methods for each protein, gene, lncRNA, and miRNA, respectively, as follows:
The parameters
The AIC method is a statistical measure used for model order selection. A lower AIC value indicates a better system model order, suggesting that the system model has achieved a good fit with fewer parameters. Therefore, we minimize the four system order detection models for each protein, gene, lncRNA, and miRNA in the candidate GWGEN using the following AIC Equations (25) to (28):
The number of nodes in candidate GWGEN and real GWGEN of FTD and healthy control
| Nodes | Candidate GWGEN | Real GWGEN of FTD | Real GWGEN of healthy control |
|---|---|---|---|
| Receptor | 1,859 | 1,859 | 1,859 |
| TF | 1,132 | 1,132 | 1,132 |
| Protein | 11,775 | 11,771 | 11,774 |
| miRNA | 150 | 150 | 150 |
| LncRNA | 196 | 187 | 189 |
| Total | 15,112 | 15,099 | 15,104 |
The number of edges in candidate GWGEN and real GWGEN of FTD and healthy control
| Nodes | Candidate GWGEN | Real GWGEN of FTD | Real GWGEN of healthy control |
|---|---|---|---|
| PPIs | 3,134,515 | 1,854,695 | 1,891,770 |
| TF-Receptor | 9,728 | 2,361 | 2,103 |
| TF-TF | 7,900 | 1,778 | 1,639 |
| TF-Protein | 57,586 | 14,085 | 12,594 |
| TF-miRNA | 450 | 82 | 77 |
| TF-LncRNA | 273 | 124 | 128 |
| miRNA-Receptor | 6,718 | 1,307 | 1,214 |
| miRNA-TF | 5,763 | 1,168 | 1,057 |
| miRNA-Protein | 40,346 | 8,141 | 7,764 |
| miRNA-miRNA | 4 | 3 | 4 |
| miRNA-LncRNA | 149 | 44 | 36 |
| LncRNA-Receptor | 163 | 34 | 47 |
| LncRNA-TF | 161 | 34 | 45 |
| LncRNA-Protein | 1,299 | 368 | 423 |
| LncRNA-miRNA | 0 | 0 | 0 |
| LncRNA-LncRNA | 3 | 0 | 1 |
| Total | 3,265,058 | 1,884,224 | 1,918,902 |
(III) Extraction of the core GWGEN by PNP method
After applying the system identification and system order detection methods, we obtained the real GWGEN for FTD and healthy control. However, the real GWGEN is still too complex to study directly. Additionally, to understand the pathogenic mechanisms of FTD, we need to use KEGG pathways for annotation. However, KEGG pathways can currently annotate only GWGENs with up to 6,000 nodes. Therefore, we use the PNP method to extract the top 6,000 nodes from the real GWGEN to form the core GWGEN for both FTD and healthy control. The PNP method involves performing singular value decomposition (SVD) on the real GWGEN. To begin, we first construct a composite network matrix A for the real GWGEN, which includes all the estimated parameters of the real GWGEN, as follows:
The following network matrix A is the expanded form of these submatrices:
Each row in A represents the interaction abilities of each protein with other proteins, or the regulatory abilities of each gene, lncRNA, and miRNA by TFs, lncRNAs, and miRNAs. Next, we perform SVD on the matrix A as follows:
Based on the SVD calculation, σr represents the r-th singular value, and σX*+Y*+Z* represents the last singular value. We select the top R singular values, which together account for at least 85% of all singular values, i.e., at least 85% of the network matrix in (30) from the network energy perspective.
In Equation (33), Er represents the proportion of the total network energy accounted for by the top r singular values. Based on this definition, we can determine the top R singular values that account for at least 85% of the total network energy.
We then project each node (i.e., wj of each row in A) of the composite network matrix A onto the top R singular vectors (i.e., vi, i = 1, …, R):
Using the PNP method, we extract the top 6,000 nodes with the highest P(wj) values from the real GWGENs of both FTD and healthy control to form the core GWGENs for both conditions, as shown in Figure S2. This is the maximum number of nodes that KEGG pathways can annotate. After KEGG annotation, we can identify the core signaling pathways for FTD and healthy control. Based on the signal transmission paths of the core signaling pathways of FTD, along with their downstream target genes and cellular dysfunctions, we investigate the pathogenic mechanisms of FTD and select the most suitable genes or proteins as significant biomarkers for FTD pathogens and drug targets for FTD treatment.
(IV) Designing a multi-molecule drug for the treatment of FTD using a DNN-based DTI model and drug design specifications
After identifying the significant biomarkers of the pathogenic mechanism of FTD, we use these biomarkers as drug targets. Next, we train a DNN-based DTI model using DTI databases to predict the interaction probabilities between drugs and targets.
First, to train the DNN-based DTI model, we integrated multiple drug-target interaction databases, including BindingDB, ChEMBL, DrugBank, PubChem, and UniProt. These databases provide information on the features of drugs and their targets, as well as the interactions between molecules. Drug features include molecular properties such as structure, topology, and geometric descriptors. Target features are described based on the physicochemical and structural properties of proteins and peptides in amino acid sequences. Using Python’s PyBioMed package, we convert the drug and target features into feature vector representations. The expression of the converted drug-target feature vectors is shown as follows:
Before using the drug-target feature vectors as training data for the DNN-based DTI model, we preprocess the feature vectors to avoid potential bias issues in the model. Specifically, the data of unverified drug-target interactions (negative class) is much larger than that of confirmed drug-target interactions (positive class). To address this, we randomly sample the unverified drug-target interactions to equalize the sample size with that of the confirmed interactions. Additionally, because the feature vector variables for drugs and targets use different units, we standardize them as follows:
Since the input layer of the DNN network used in the DTI model has only 996 nodes, while the standardized feature vectors in Equation (35) still exceed this number, we use principal component analysis to reduce the dimensionality of the drug-target feature vectors. After reducing the dimensionality of the drug-target feature vectors to 996, we use 75% of the drug-target feature vector data as training data and the remaining 25% as testing data. We then use Python’s TensorFlow and Keras libraries for training and prediction. The architecture of the DNN includes four hidden layers, both in the input and output layers, with the hidden layers employing the rectified linear unit activation function. Dropout layers are added to each hidden layer to avoid overfitting. The output layer uses a sigmoid activation function to constrain the output between 0 and 1, representing the probability of interaction between the drug and the target. The neural network parameters are set with a learning rate of 0.001, 100 epochs, and a batch size of 100. Additionally, the Adam optimization algorithm is used for training the neural network. Since drug-target interaction is a binary classification problem (interaction or no interaction), we use binary cross-entropy as our loss function:
To optimize the weight vector w and bias vector b, we combine them into a vector θ and use the backpropagation algorithm to compute the gradient and obtain the optimal model parameter set θ*. The advantage of the backpropagation algorithm is that it can efficiently compute high-dimensional vectors and adjust the DTI model parameters to fit the drug-target interaction data for each iteration. The gradient iteration algorithm is as follows:
To evaluate the performance of our trained DTI model, we use the five-fold cross-validation method. We divide the training data into five equal parts and, in each iteration, use one part as the validation data and the remaining four parts as the training data. We average the evaluation results of the five validations to obtain the final performance metric of the model, as shown in Figures S3 and S4. Additionally, we use the area under the curve (AUC) of the receiver operating characteristic (ROC) curve as another reference metric. For binary classification problems, AUC is an important indicator for evaluating the performance of the model visually. The larger the AUC, the better the performance of the DNN-based DTI model. The AUC equations for the ROC curve are given as follows:
After predicting the interaction probabilities between drugs and targets, we also need to consider drug design specifications to further refine the candidate drugs and select a suitable multi-molecular drug for treating FTD. We use three drug design specifications for screening: regulatory capacity, sensitivity, and toxicity, as shown in Table S1. For regulatory capacity, we used the LINCS L1000 Level 5 database, where a regulatory ability >0 indicates upregulation of expression levels, and <0 indicates downregulation. Sensitivity was assessed using the PRISM repurposing dataset, representing the compound’s interference with human cells, with values closer to 0 indicating lower interference. Toxicity was evaluated using the ADMETlab 2.0 tool, where the standardized value LC50 indicates toxicity, with higher values representing lower toxicity to the human body. Based on strong regulatory capacity, high sensitivity, and low toxicity, we propose potential molecular drug combinations as candidates for multi-molecular drugs to treat FTD.12
The other detailed methods are provided in the Supplementary materials.
Results
Investigation of core signaling pathways using systems biology methods and prediction of candidate drugs using a trained DNN-based DTI model by DTI databases
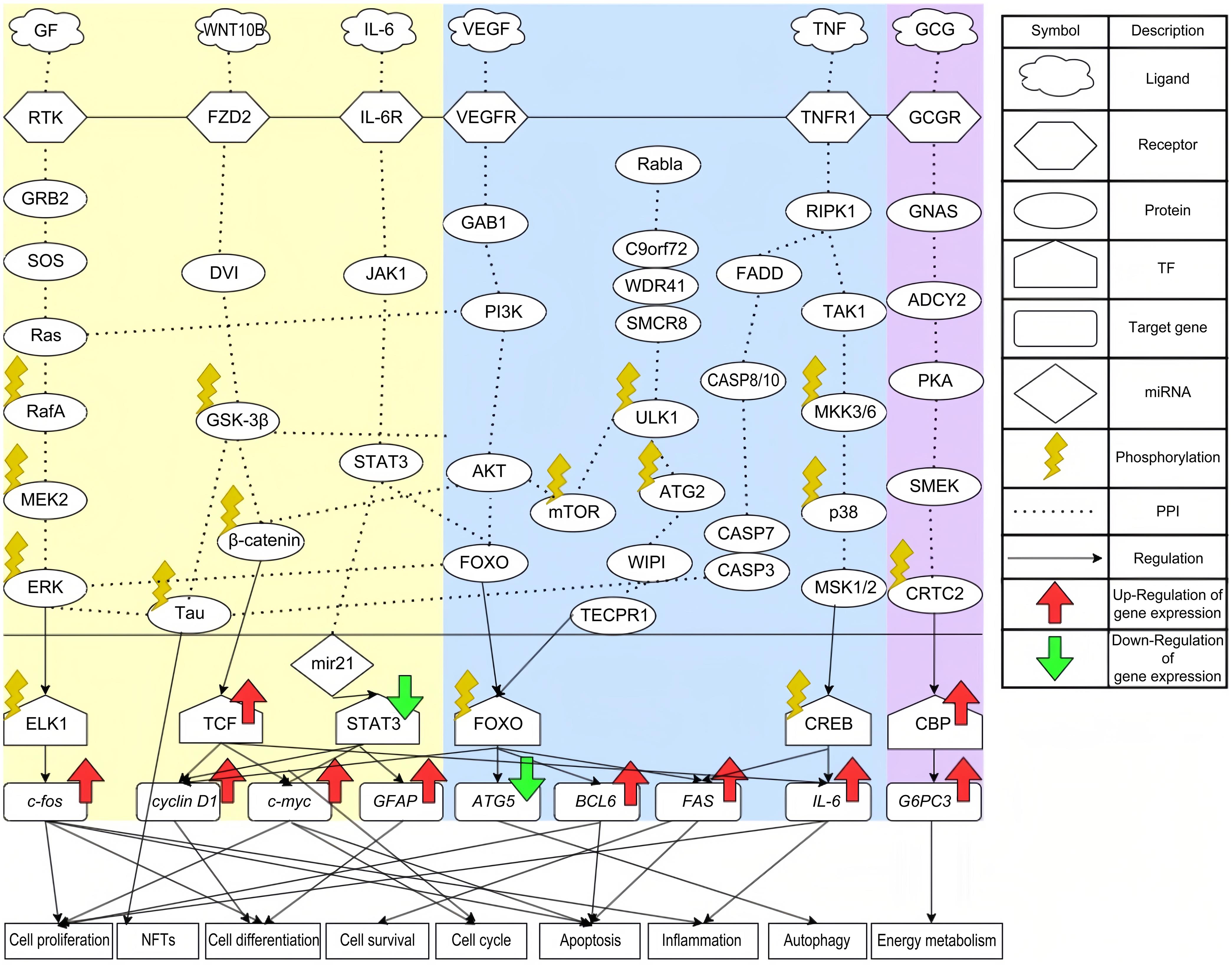
Following KEGG pathway annotation, the core signaling pathways for FTD (as presented in Table 3) and healthy controls (as presented in Table 4) are illustrated in Figure 2. In the following subsection, we will analyze cytokines in the microenvironment, core signaling pathways, their downstream target genes, and associated cellular dysfunctions to explore the pathogenic mechanisms of FTD. We will then select biomarkers that play a key role in the pathogenesis of FTD, such as TAU, GSK-3β, STAT3, ATG5, WDR41, and RIPK1. By applying the DNN-based DTI model, trained on the DTI database, we predict potential drugs and select candidate molecular drugs based on design specifications to formulate a multi-molecule drug aimed at treating FTD by targeting these significant biomarkers.
Core signaling pathways of FTD through annotation of KEGG pathways
| Pathway | Gene number | p-value |
|---|---|---|
| MAPK signaling pathway | 148 | 2.4E-10 |
| WNT signaling pathway | 76 | 1.1E-3 |
| JAK-STAT signaling pathway | 72 | 2.6E-3 |
| PI3K-Akt signaling pathway | 135 | 1.9E-2 |
| Amyotrophic lateral sclerosis | 166 | 1.3E-7 |
| TNF signaling pathway | 69 | 8.8E-9 |
Core signaling pathways of healthy control through annotation of KEGG pathways
| Pathway | Gene number | p-value |
|---|---|---|
| Cell Cycle | 86 | 1.2E-9 |
| PI3K-Akt signaling pathway | 141 | 7.0E-4 |
| Amyotrophic lateral sclerosis | 148 | 1.4E-4 |
| TNF signaling pathway | 56 | 2.7E-4 |
| Apoptosis | 65 | 4.6E-5 |
| Nucleocytoplasmic transport | 60 | 2.0E-7 |
The left area of this picture presents the core signaling pathways of FTD. The middle area shows the common core signaling pathways between FTD and healthy control. The right area presents the core signaling pathways of healthy control. The green and red arrows represent abnormal downregulation and upregulation of selected significant biomarkers of FTD, respectively. TF, Transcription Factor; PPI, Protein-Protein Interaction; NFT, Neurofibrillary Tangle.
The role of biomarker TAU in the MAPK signaling pathway
The growth factor (GF) family comprises proteins or peptides that control diverse cellular functions, including cell growth, differentiation, and survival. They play crucial roles in processes like development, tissue repair, immune response, and intercellular communication.13 Receptor tyrosine kinases serve as receptors for GFs and are essential for neuronal function and development, as shown in Figure 2. For instance, neurotrophins and other GFs, which are expressed in very limited amounts, play a vital role in regulating neuronal development, plasticity, and survival.14 Receptor tyrosine kinases, upon receiving signals from GFs, activate downstream signaling pathways, such as the GRB2/SOS/Ras signaling pathway. The activation of Ras is essential for processes such as cell proliferation, differentiation, and apoptosis.15 RafA is a serine protein kinase that, upon receiving signals from Ras, can directly phosphorylate proteins or activate downstream MEK/ERK pathways to promote protein phosphorylation and regulate cell apoptosis.16 ERK, a member of the MAPK family, is responsible for phosphorylating the transcription factor ELK1, which upregulates c-fos. The ERK/ELK1/c-fos pathway is involved in inflammatory responses, cell differentiation, cell proliferation, and apoptosis.17 Additionally, the persistent activation of ERK induces the phosphorylation of TAU protein. Various studies have indicated that TAU phosphorylation causes neurofibrillary tangles (NFTs),18 one of the most common initial symptoms of FTD.
The role of biomarker GSK-3β in the WNT signaling pathway
Research has shown that the WNT signaling pathway can lead to mutations in progranulin, which are a significant pathogenic mechanism in FTD,19 as shown in Figure 2. FZD2, serving as a receptor in the WNT signaling pathway, plays a crucial role in this process. A decrease in FZD2 levels results in increased cell apoptosis, while its upregulation promotes the survival of neurons in vitro.20 After receiving the WNT signal, FZD2 activates GSK-3β, an important kinase for TAU. As discussed in the previous section, the phosphorylation of TAU plays a potential pathogenic role in FTD. In addition to causing TAU phosphorylation, GSK-3β is essential in the WNT pathway. It phosphorylates downstream β-catenin, leading to its degradation through the ubiquitin-proteasome pathway. β-catenin serves as an activator of T-cell factor-dependent transcription, resulting in the upregulation of various target genes, such as c-myc and cyclin D1.21 The expression of c-myc is strongly linked to cell cycle progression and can also trigger apoptosis. As the cell cycle initiates, cyclin D1 moves through the entire cycle,22 regulating cell proliferation and participating in cell differentiation.
The role of biomarker STAT3 in the JAK-STAT signaling pathway
Interleukin-6 (IL-6) is produced by keratinocytes and white blood cells, and since its discovery, the IL-6 signaling pathway has become a core pathway involved in healthy immune regulation and immune dysregulation in many diseases. Research has found that elevated levels of IL-6 can contribute to granulin precursor mutations, which are also a key pathogenic mechanism of FTD.23 IL-6R receives IL-6 and activates JAK1 and STAT3, as shown in Figure 2. The JAK/STAT signaling pathway coordinates adaptive and innate immune mechanisms, ultimately limiting neuroinflammatory responses and acting as a key contributor to neuroinflammation in neurodegenerative diseases.24 The downstream effector miR-21 of STAT3 is a miRNA associated with dysfunction in neuron-glial cell function. Some studies have shown it to be overexpressed in neurons derived from induced pluripotent stem cells of FTD patients with the PSEN1ΔE9 deletion (iNEU-PSEN).25 Additionally, miR-21 is linked to the regulation of toxicity caused by amyloid-beta (Aβ) oligomers, which is generally considered one of the pathological mechanisms underlying neurodegenerative diseases in the brain. Finally, miR-21 activates the transcription factor STAT3, which upregulates many target genes, including c-myc, cyclin D1, and GFAP. The functions of c-myc and cyclin D1 were introduced in previous sections. As for GFAP, it is a glial fibrillary acidic protein found in astrocytes in the central nervous system, where it plays a role in cell differentiation. Several studies have indicated that serum GFAP levels are significantly higher in FTD patients than in healthy controls.26
The role of biomarker ATG5 in the phosphoinositide 3-kinase (PI3K)-Akt signaling pathway
Vascular endothelial growth factor (VEGF) is involved in neurodevelopment, angiogenesis, and hematopoiesis, playing an essential role in maintaining homeostasis in the adult vascular system. A study reported that elevated levels of VEGF can cause hippocampal atrophy. Over time, hippocampal atrophy can lead to cognitive decline, which may contribute to the development of FTD,27 as shown in Figure 2. VEGFR receives the VEGF signal and transmits it through GRB2-associated binding protein 1 to the downstream PI3K/AKT pathway.28 The PI3K and protein kinase B (AKT) signaling pathway play roles in many important cellular functions. In the brain, the PI3K/AKT signaling pathway serves various functions, including regulating survival, cell proliferation, growth, differentiation, and other complex processes. It also plays a role in oxidative stress and autophagy during neuroinflammation.29 The PI3K/AKT pathway phosphorylates downstream GSK-3β, which, as mentioned earlier, contributes to neurofibrillary tangle formation. In addition to its impact on GSK-3β, AKT can also phosphorylate downstream proteins mTOR and FOXO. Phosphorylation of FOXO by AKT inhibits FOXO’s transcriptional function, promoting cell survival and proliferation. The transcription factor FOXO targets many genes, including cyclin D1, ATG5, BCL6, and FAS. FOXO induces apoptosis by upregulating mitochondrial-targeting proteins of the Bcl family.30ATG5 is a gene involved in the autophagy process and also participates in regulating cell survival and metabolic balance, which is crucial for maintaining cellular function. However, FOXO downregulates ATG5, leading to changes in the autophagy process. A lack of autophagy can impair learning and memory, which is one of the important symptoms in patients with FTD.31
The role of biomarker WDR41 in the amyotrophic lateral sclerosis signaling pathway
The GGGGCC hexanucleotide repeat expansion in the C9orf72 gene is the leading genetic cause of amyotrophic lateral sclerosis and FTD.32 In related studies, we have found that C9orf72, along with SMCR8 and WDR41, forms a stable complex through their interaction, which is involved in the regulation of macroautophagy. The C9orf72-SMCR8-WDR41 complex interacts with the autophagy initiation complex involving Rab1a and Unc-51-like kinase 1 (ULK1).33 As an effector of Rab1a, the C9orf72-SMCR8-WDR41 complex regulates the initiation of autophagy by controlling Rab1a-dependent transport of the ULK1 autophagy initiation complex to phagophores. Within the C9orf72-SMCR8-WDR41 complex, WDR41 is a prominent C9orf72-interacting protein, playing a significant role in supporting the regulatory association of C9orf72 with lysosomes. In the complex, SMCR8 acts as an upstream component of ULK1. The interaction between ULK1 and mTOR is essential for autophagic function. After the fusion of the ULK1 autophagosome with the lysosome, mTOR can be reactivated. The activation of mTOR subsequently reduces ULK1 kinase activity by phosphorylating it at the Ser757 site, thereby suppressing autophagy.34 The phosphorylation of ULK1 also leads to the phosphorylation of its downstream protein Atg2. Atg2, as a key protein in membrane expansion during the ULK1-initiated autophagy process, ensures the formation of autophagosomes.35 By binding to WIPI proteins, Atg2 is localized and stabilized on the autophagosome membrane, promoting the transport of membrane lipids and membrane expansion.36 Another role of WIPI is to activate TECPR1, which transmits signals to the transcription factor FOXO to downregulate ATG5. This downregulation results in the impairment of autophagic function, contributing to neurodegenerative diseases.
The role of biomarker RIPK1 in the tumor necrosis factor (TNF) signaling pathway.
The primary pro-inflammatory cytokine TNF has been demonstrated to regulate various signaling pathways, leading to a broad range of downstream effects.37 These effects encompass the regulation of cell proliferation, differentiation, apoptosis, immune response, and the induction of inflammation. Due to such extensive cellular effects and complex signaling pathways, TNF is also associated with many age-related disease states. Upon receiving the TNF signal, the receptor TNFR1 begins to activate downstream pathways.37 The RIPK1 is recognized as a key regulator of TNFR1 signal transduction. RIPK1 controls cell fate decisions, promoting either cell survival or death. The downstream FADD/CASP signaling pathway is responsible for regulating extrinsic apoptosis and necroptosis.38 Caspase-3 (CASP3) in the CASP family can cleave TAU protein, leading to its phosphorylation and the formation of NFTs.39 In addition to regulating cellular apoptosis, RIPK1 is also involved in mediating inflammatory responses in neurodegenerative diseases. The downstream TAK1/MKK/p38 signaling pathway plays an essential role in inflammation, as activation of p38 induces the expression of inflammatory mediators involved in tissue remodeling and oxidative regulation.40 Downstream of p38, mitogen- and stress-activated protein kinases 1 and 2 act as epigenetic modifiers that activate genes related to cell proliferation, inflammation, and neuronal function, as well as phosphorylate the transcription factor CREB.41 CREB has a vital role in the nervous system, participating in the formation of learning and memory, and is responsible for upregulating target gene IL-6. IL-6 is important for neuronal development, differentiation, and regeneration; therefore, its dysregulation is associated with neuroinflammation to some degree.42
The core signaling pathways of healthy control
In the healthy control tissues, we observed that the glucagon signaling pathway plays a crucial role. However, recent studies indicate that, besides its role in glucose metabolism, the glucagon signaling pathway also protects the nervous system by regulating neuronal metabolism, antioxidant stress response, and inflammation.43 Upon binding of pancreatic glucagon and its receptor in the microenvironment, the GNAS/ADCY2/PKA/SMEK signaling pathway is activated, as shown in Figure 2. The actions of these kinases are also linked to metabolic regulation. Overexpression of SMEK, the downstream component, leads to phosphorylation of CRTC2.44 Phosphorylated CRTC2 then interacts with CREB-binding protein, regulating the expression of the target gene G6PC3. The main function of G6PC3 is to maintain energy homeostasis and mitochondrial function. In this study, we observed that the expression level of the G6PC3 gene is significantly elevated in normal tissues compared to those in FTD patients, which may explain the metabolic abnormalities in FTD patients due to G6PC3 deficiency.45
Predicting potential drugs for treating FTD using biomarkers as drug targets and leveraging a deep neural network-based drug-target interaction model
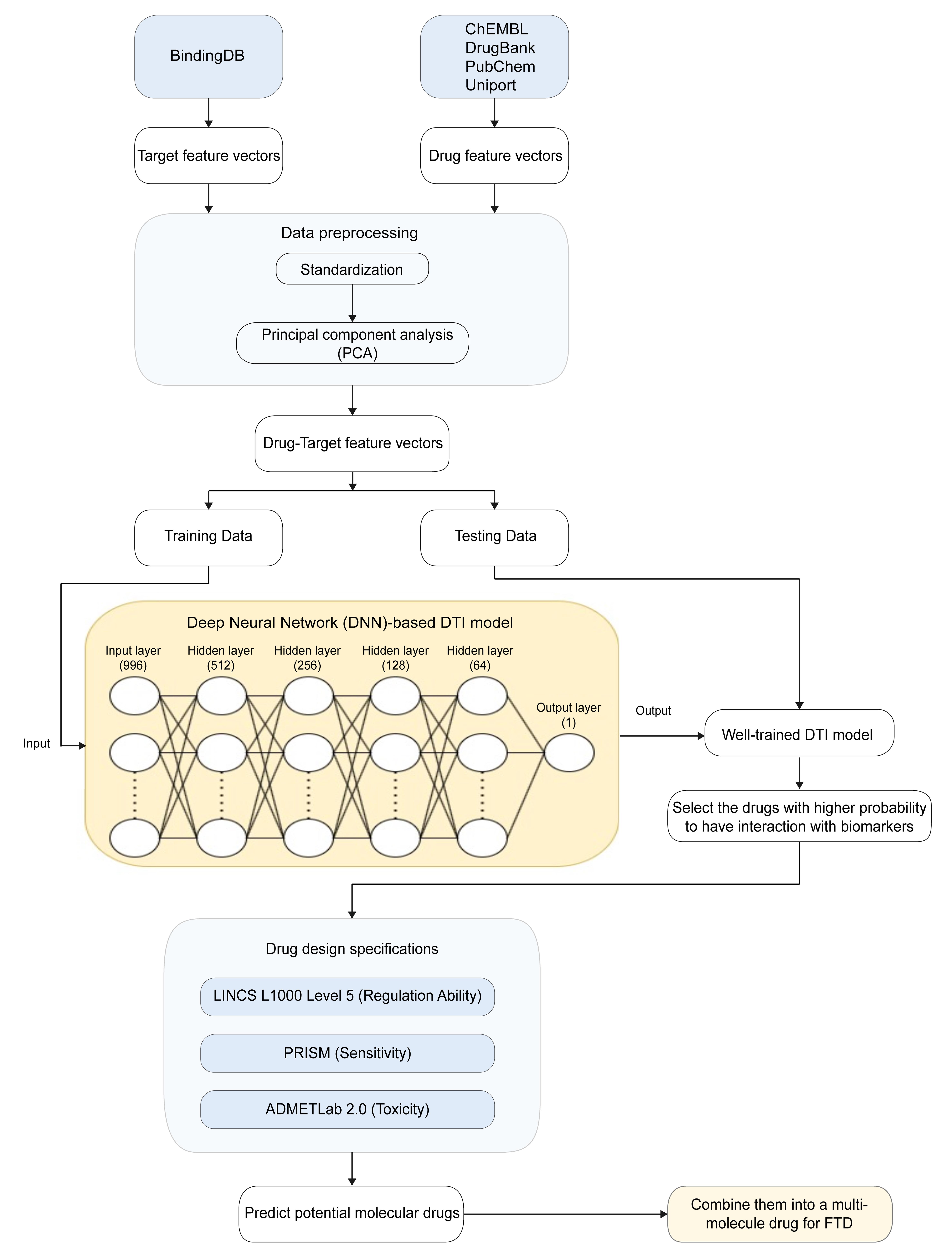
After investigating the core signaling pathways involved in the pathogenic mechanism of FTD and identifying significant biomarkers TAU, GSK-3β, STAT3, ATG5, WDR41, and RIPK1 as drug targets, we began studying the interactions between these biomarkers and drugs, considering drug design specifications such as regulatory ability, sensitivity, and toxicity. Based on these significant drug properties, we selected potential drugs expected to reverse the expression levels of these biomarkers without causing excessive side effects. To study the interactions between biomarkers and drugs, we developed a DNN-DTI model, as shown in Figure 3. This model was pre-trained using the DTI database, enabling it to effectively predict the interaction probabilities between biomarkers and candidate drugs after DTI data training via the Adam learning algorithm.
First, we preprocess the original data through drug-target interaction databases to generate the drug-target feature vectors. Seventy-five percent of the drug-target feature vector data was used as training data, and the remaining 25% was used for testing. After training the DTI model, we obtain the score for each molecular drug (i.e., the higher the score, the higher the probability of interaction with biomarkers). We then select candidate molecular drugs according to their scores. Finally, we use three drug design specifications to predict the most promising molecular drugs and combine them into a multi-molecule drug for treating FTD.
However, in the DTI database, there were 80,291 confirmed drug-target interactions but 100,024 unconfirmed interactions. To address the prediction issues caused by the imbalanced class distribution, we randomly selected 80,291 unconfirmed drug-target interactions for prediction. Another potential issue arises from the need to observe multiple variables in drug-target interaction feature data, with many variables possibly correlated, increasing the complexity of the analysis. Therefore, we standardized the data and used principal component analysis to reduce the dimensionality of the feature vectors to 996, in order to meet the input requirements of the DNN for computational convenience. These 996 nodes were used as the input layer, with the DNN-based DTI model comprising four hidden layers with 512, 256, 128, and 64 neurons, respectively. The hidden layers employed rectified linear unit activation functions, and dropout layers were added to each hidden layer to avoid overfitting. The output layer consisted of a single neuron using a sigmoid activation function to constrain the output between 0 and 1, representing the interaction probability between the drug and the drug target.
Our DNN-based DTI model was evaluated using 5-fold cross-validation to assess its performance. The loss and accuracy curves during the training process are shown in Figures S3 and S4 of the supplementary materials, respectively, and the results of the five-fold cross-validation are presented in Table S2 of the supplementary materials. The average test accuracy was 93.2%, with a standard deviation of 0.118%. Additionally, we used ROC curves as a reference metric, where the AUC of the ROC ranges between 0 and 1, with higher values indicating better DTI model performance. Ultimately, our DNN-based DTI model achieved an AUC of 0.981, as shown in Figure S5 of the supplementary materials, outperforming a random prediction model (AUC = 0.5).
However, we aimed to design molecular drugs that not only effectively alleviate symptoms of FTD but also minimize adverse reactions. Generally, potent molecular drugs may be more irritating to the human body. Therefore, balancing drug efficacy and side effects is our next focus. We used three drug design specifications for screening the predicted molecular drugs: Regulatory capacity, sensitivity, and toxicity, referring to the LINCS L1000 Level 5, PRISM, and ADMETlab 2.0 databases. Based on strong regulatory capacity, high sensitivity, and low toxicity, we selected several potential molecular drugs for each biomarker of FTD, as shown in Table 5. Additionally, since Iodophenpropit, TTNPB, Probucol, and Clobenpropit serve as potential molecular drugs for these significant biomarkers, we combined these four molecular drugs to propose potential multi-molecular drugs for treating FTD.
Using drug design specifications to select potential molecule drugs
| Drug | Target | |||||||
|---|---|---|---|---|---|---|---|---|
| TAU | GSK-3β | STAT3 | ATG5 | WDR41 | RIPK1 | Sensitivity (PRISM) | Toxicity (LC50) | |
| Iodophenpropit | ↑ | ↓ | ↓ | −0.0032 | 4.94 | |||
| Clobenpropit | ↓ | ↓ | ↑ | 0.0416 | 4.223 | |||
| TTNPB | ↓ | ↓ | ↓ | 0.2480 | 5.823 | |||
| Probucol | ↓ | ↓ | ↓ | −0.2931 | 7.007 | |||
Discussion
Iodophenpropit and Clobenpropit are both antagonists of H3 receptors commonly used to study the role of histamine in the nervous system. The histamine H3 receptor is a biogenic amine that plays a significant role in central nervous system activities, including learning and memory. Literature data indicate that elevated levels of H3 receptor expression can lead to cognitive impairment.46 Given preclinical evidence that blocking the H3 receptor reduces impulsivity, improves attention, and enhances learning and memory, H3 receptor antagonists have been clinically used to treat various cognitive disorders. Iodophenpropit, as one of these H3 receptor antagonists, was the first compound successfully developed for labeling H3 receptors in rat brain membranes in previous studies.46 In another study, Clobenpropit was found to improve memory impairment induced by lipopolysaccharides. Lipopolysaccharide triggers neuroinflammation by modulating cyclooxygenase activity and cytokine levels in the brain, leading to memory deficits.47
TTNPB is a synthetic analog of all-trans retinoic acid, belonging to the vitamin A derivatives (retinoid) family. Retinoids interact with retinoic acid receptors and retinoid X receptors, significantly impacting physiological and pathological signaling pathways in the brain. Impaired retinoic acid signaling is a crucial factor leading to neurodegenerative diseases.48 Recent studies have shown that deprivation of retinoic acid in mice results in severe deficits in spatial learning and memory. Additionally, a critical pathological hallmark of retinoids is the production and deposition of Aβ, the formation and phosphorylation of NFT, and the inflammation and autoimmune response.49 These pathologies are also significant mechanisms in the etiology of FTD. Currently, TTNPB has been identified in clinical settings as a potent retinoid receptor agonist with potential for treating neurodegenerative diseases due to its metabolic resistance and high affinity for retinoic acid receptors.48
Probucol is a historically established cholesterol-lowering drug, but recent studies have explored its potential as a treatment for dementia. This interest arises because Probucol has been shown to inhibit Aβ secretion in mouse models while maintaining blood-brain barrier function, suppressing neurovascular inflammation, and directly influencing neuroprotection and adaptability.50 In mouse models with ischemia-induced blood-brain barrier dysfunction, Probucol preserved the proper localization of tight junction proteins in endothelial cells by attenuating sphingosine-1-phosphate signaling and inhibiting the expression of STAT3, thereby reducing the leakage of small molecules into the brain parenchyma. Furthermore, in in vitro models of brain endothelial dysfunction, Probucol was found to inhibit the expression of CASP3.50 The cleavage of CASP3 is one of the causes of TAU phosphorylation, and inhibiting CASP3 can reduce the formation of NFT caused by TAU phosphorylation. Taken together, these findings suggest that Probucol may provide therapeutic benefits for FTD by effectively enhancing neuronal survival and plasticity, making it a promising candidate for FTD treatment.51
In summary, there is currently no drug on the market that can completely cure FTD. The four molecular drugs we have screened have not yet been practically applied to treat FTD in humans. However, based on our systematic research and analysis, by understanding the mechanisms of action of these molecular drugs, effectively regulating the expression of biomarkers, and improving the pathogenic mechanisms of FTD, we have ultimately selected these four molecular drugs as potential treatments for FTD. Compared to traditional drugs, these small-molecule compounds we have chosen as a molecular drug combination offer several advantages. Firstly, development costs: Traditional drugs and multi-molecular drugs are associated with high costs, time consumption, and low efficiency. Secondly, multi-target action: Multi-molecular drugs can act on multiple targets simultaneously, thereby increasing efficacy, especially for complex diseases like neurodegenerative disorders.
Conclusions
In this study, we explored the pathogenic mechanism of FTD from a systems biology perspective and designed a multi-molecule drug for its treatment. To achieve this goal, we began by constructing candidate GWGENs for FTD and healthy control through big data mining, including candidate PPINs and candidate GRNs. The next step was to identify the true GWGENs for FTD and healthy control using their microarray data and system identification methods. Subsequently, we applied the PNP method to extract the core GWGENs for both FTD and healthy control. By annotating these core GWGENs with KEGG pathways, we identified the core signaling pathways involved in FTD and healthy control and investigated the pathogenetic mechanisms to pinpoint significant biomarkers for FTD.
In the core pathogenic signaling pathways of FTD, we identified significant biomarkers, including TAU, GSK-3β, STAT3, ATG5, WDR41, and RIPK1, as potential drug targets. Based on the prediction of candidate molecular drugs from the DNN-based DTI model, we selected Iodophenpropit, TTNPB, Probucol, and Clobenpropit as a multi-molecule drug combination targeting multiple biomarkers to restore the pathogenic cellular functions of FTD to normal levels. With further clinical and experimental validation, we hope that the proposed multi-molecule drug will improve the cellular functions in FTD patients. The potential therapeutic efficacy of these molecular drug combinations is expected to offer new treatment options for FTD patients.
Supporting information
Supplementary material for this article is available at https://doi.org/10.61474/ncs.2024.00043 .
Fig. S1
(a) The visualized graph of real GWGEN of FTD; (b) The visualized graph of Real GWGEN of healthy control. The red lines are gene regulations, and the blue lines are protein-protein interactions. The numbers present the total nodes.
(TIF)
Fig. S2
(a) The visualized graph of core GWGEN of FTD; (b) The visualized graph of core GWGEN of healthy control. The red lines are gene regulations, and the blue lines are protein-protein interactions. The numbers present the total nodes.
(TIF)
Fig. S3
The line chart of the training and validation accuracy by using 5-fold cross validation. “-o-” line denotes the training accuracy. “-◊-” line denotes the validation accuracy.
(TIF)
Fig. S4
The line chart of the training and validation loss by using 5-fold cross validation. “-o-” line denotes the training loss. “-◊-” line denotes the validation loss.
(TIF)
Fig. S5
The area under curve (AUC) of the Receiver Operating Characteristic (ROC). AUC is an important indicator for evaluating the performance of the model visually. The larger the AUC, the better the performance of the DNN-based DTI model. The AUC value of our model is 0.981.
(TIF)
Table S1
Using drug design specifications to select candidate molecule drugs for each biomarker of FTD. (+) denotes overexpression; (-) denotes low expression in FTD.
(DOCX)
Table S2
The prediction performance of the DNN-based DTI model using 5-fold cross-validation (early stopping at epoch 74).
(DOCX)
Declarations
Acknowledgement
The authors thank Prof. Yung-Jen Chuang from the Institute of Bioinformatics and Structural Biology, National Tsing Hua University, Hsinchu, Taiwan for general discussions about this topic.
Ethical statement
Ethical approval is not applicable due to the use of the publicly available dataset GSE140830 (
Data sharing statement
The whole blood data for FTD and healthy controls is from GSE140830 (
Funding
None.
Conflict of interest
The authors declare no conflict of interest.
Authors’ contributions
Conceptualization (BSC), methodology (WLC), software (WLC), validation (WLC), formal analysis (WLC), investigation (WLC), data curation (WLC), writing—original draft preparation (WLC), writing—review and editing (BSC), visualization (WLC), and supervision (BSC). All authors have read and agreed to the published version of the manuscript.

 Author information
Author information