Introduction
Environmental pollutants have become a significant concern in modern society due to their detrimental effects on human health, particularly on the neurological system. The intricate interplay between environmental toxins and neurological disorders has garnered increasing attention from researchers and policymakers alike.1,2 From heavy metals and pesticides to air and water pollutants, these substances infiltrate our environment, posing a myriad of risks to neurological function.3 Understanding the mechanisms by which environmental pollutants induce neurological diseases is paramount for developing effective mitigation strategies and safeguarding public health.1,2 In this introduction, we delved into the complex relationship between environmental contaminants and neurological illness, exploring key pollutants, their routes of exposure, and the neurological diseases they can precipitate. By elucidating these connections, we aimed to raise awareness and contribute to the collective effort to address this pressing global health challenge.
In the quest for progress and convenience, humanity has harnessed the power of an extensive array of chemicals to manufacture essential substances across various sectors.4 However, this reliance on chemical compounds often comes at a significant cost to the environment, with many of these substances proving to be harmful pollutants that contribute to ecological degradation.1,5 Moreover, their impact extends beyond environmental concerns, as they are increasingly linked to the onset and exacerbation of various diseases. Chemicals used in manufacturing essential substances serve crucial functions in modern society but also pose significant risks to both environmental integrity and human health.6,7 Take, for instance, pesticides sprayed on crops to enhance agricultural yields. While these chemicals help combat pests and ensure food security, their residues can contaminate soil and water, leading to ecological imbalances and potentially exposing humans to harmful toxins linked to neurological disorders,8 developmental delays, and even certain types of cancer.9,10
Cadmium acetate, a chemical compound with the formula Cd(C2H3O2)2, has several applications in various industries, such as electroplating, chemical synthesis, and the textile industry as a mordant, despite its toxicity. Exposure to cadmium (Cd) in the environment can lead to its accumulation in the body, including the brain, through various routes of contact. Once inside the central nervous system, cadmium disrupts neuronal function and signaling pathways, leading to oxidative stress, inflammation, and neuronal damage. This can result in a range of neurological symptoms and disorders, including cognitive deficits, motor dysfunction, and neurodegenerative diseases such as Parkinson’s and Alzheimer’s. Various antioxidants and natural substances may serve as potent therapeutic agents, as evidenced in different studies.11–14
Alzheimer’s disease (AD) is a complex neurodegenerative disorder characterized by the accumulation of abnormal protein aggregates, including beta-amyloid plaques and tau tangles, in the brain. Glycogen synthase kinase-3 beta (GSK-3β), a multifunctional serine/threonine kinase, has been implicated in the pathogenesis of AD through various mechanisms:
Tau Phosphorylation: GSK-3β plays a crucial role in the hyperphosphorylation of tau protein, a microtubule-associated protein essential for neuronal structure and function.
15 ,16 Hyperphosphorylated tau loses its ability to stabilize microtubules, leading to the formation of neurofibrillary tangles, a hallmark of AD pathology.17 Beta-Amyloid Production: GSK-3β influences the processing of amyloid precursor protein into beta-amyloid peptides, a key component of senile plaques in AD brains. GSK-3β promotes the cleavage of amyloid precursor protein by β-secretase and γ-secretase, leading to the generation of toxic beta-amyloid peptides that aggregate and contribute to neuronal dysfunction and cell death.
18 ,19 Neuronal Apoptosis: GSK-3β activation can trigger apoptotic pathways in neurons, leading to cell death. This process may be mediated by various mechanisms, including mitochondrial dysfunction, oxidative stress, and dysregulation of pro-survival signaling pathways.
20 Synaptic Dysfunction: GSK-3β dysregulation has been implicated in synaptic impairment, including synaptic loss and synaptic plasticity deficits, which are early events in AD pathogenesis. GSK-3β-mediated disruption of synaptic function contributes to cognitive decline and memory impairment characteristic of AD.
21 ,22 Neuroinflammation: GSK-3β activation contributes to neuroinflammatory processes observed in AD brains, including activation of microglia and astrocytes, release of pro-inflammatory cytokines, and induction of neuroinflammatory signaling cascades, further exacerbating neuronal damage and dysfunction.
23 ,24 Overall, GSK-3β dysregulation plays a multifaceted role in the pathogenesis of Alzheimer’s disease, contributing to tau pathology, beta-amyloid accumulation, neuronal apoptosis, synaptic dysfunction, and neuroinflammation.
Cd induces dysregulation of GSK-3β through several interconnected mechanisms.25 As a toxic heavy metal, Cd can activate GSK-3β directly or indirectly by disrupting cellular signaling pathways that regulate its activity, resulting in increased GSK-3β activity.26 Once activated, GSK-3β excessively phosphorylates tau protein, leading to the formation of neurofibrillary tangles characteristic of AD.16 Additionally, Cd-induced GSK-3β activation influences the processing of amyloid precursor protein, contributing to increased production of toxic beta-amyloid peptides and the formation of senile plaques.27 Furthermore, GSK-3β activation by Cd triggers neuronal apoptosis through various mechanisms, including mitochondrial dysfunction and oxidative stress, ultimately leading to neurodegeneration.26,28 Moreover, dysregulation of GSK-3β by cadmium may impair synaptic function, contributing to synaptic loss and cognitive decline in AD.16 Understanding these mechanisms is essential for developing targeted therapeutic interventions to counteract the toxic effects of cadmium and alleviate AD progression.
This research aimed to fill gaps in understanding how Cd induces dysregulation of GSK-3β. Through in-silico methods, we aimed to explore the molecular mechanisms involved. This includes utilizing computational modeling to uncover how Cd affects GSK-3β activity, investigating structural changes in GSK-3β due to Cd exposure, predicting binding sites between Cd and GSK-3β, and identifying potential therapeutic targets to mitigate neurological effects. While previous studies have linked cadmium toxicity to neurodegeneration, this work specifically investigates cadmium acetate’s direct interaction with GSK-3β at the molecular level, providing new insights into its role in Alzheimer’s disease progression. Unlike prior studies that focused on general toxicity mechanisms, this research identifies specific binding residues and their implications for enzyme dysregulation. Ultimately, this study seeks to enhance our understanding of Cd-induced GSK-3β dysregulation and its potential for treating associated neurological disorders like AD.
Materials and methods
Sequence retrieval
The sequence and structure of GSK-3β related to neurotoxicity were obtained from the Protein Data Bank (PDB) with the PDB ID 1i09. It consists of two chains, namely chain A and chain B. The receptor protein was prepared by eliminating the ligand and water molecules from the active site and introducing polar hydrogens.
Compound preparation
The 3D conformer of cadmium acetate [Cd(C2H3O2)2], with a PubChem compound identifier of 10986 and a molecular weight of 230.50 g/mol, was obtained from PubChem (pubchem.ncbi.nlm.nih.gov) and utilized as the ligand in this study.
Selection of binding site
Blind docking of protein-ligand complexes offers a robust approach for exploring receptor binding sites and the distinct poses adopted by ligands. Its versatility has led to its extensive application in pharmaceutical and biological research fields.29 Due to the limited literature on the ligand binding site of the human GSK-3β enzyme, blind docking was chosen as the methodology for this study.
Molecular docking
In the molecular docking process, the methodology established in our prior research was applied.30 Molecular docking was conducted using GSK-3β (PDB ID 1i09). In this study, rigid docking was employed, as indicated by the preparation of the receptor protein (GSK-3β), which involved removing water molecules, adding polar hydrogens, and introducing charges. Notably, no flexibility was incorporated into either the receptor or the ligand during the docking process. This method was selected for its computational efficiency and its ability to predict binding affinities within a stable receptor conformation.31 The docking was conducted by AutoDock Vina (version 1.1.2) with an exhaustiveness parameter set to 8. The grid box dimensions were set to 60 × 60 × 60 Å, centered on the GSK-3β catalytic domain. These parameters were chosen to balance computational efficiency with accuracy.32 Additionally, blind docking was utilized to explore potential allosteric binding sites beyond the active site. This method ensures unbiased identification of interaction hotspots, particularly for small molecules like cadmium acetate.33
Preparation of the macromolecular file involved the addition of polar hydrogens and the introduction of charges via Apparent Diffusion Tensor (ADT) with default Kollman charges, automatically incorporating Kollman charges for a peptide. Protein parameters were integrated, and files were saved as 1i09.pdbqt. The ligand, cadmium acetate Cd(C2H3O2)2, was converted to PDB format using Open Babel 2.3.2a, loaded into ADT, and configured accordingly, resulting in ligand.pdbqt. A grid with sufficient space for unrestricted ligand rotation was created, and parameters were stored in molecule.gpf, with a new file, 1alu.gpf, being saved. AutoGrid4 generated maps, producing 1i09.glg during the run. Docking Parameter File creation involved reading macromolecular pdbqs and ligand.out.pdbq files. AutoDock, utilizing the Lamarckian genetic algorithm, initiated the docking task. Terminal access to .dlg files recorded final docked energy, Gibbs free energy, and inhibition constant for each of the 500 runs, ensuring optimal results.
Comparative analysis with known modulators
To contextualize cadmium acetate’s interaction with GSK-3β, we compared its binding affinities and interaction profiles to those of established inhibitors such as Tideglusib and lithium chloride. The results indicate that cadmium acetate shows weaker binding affinity but targets unique residues, suggesting an indirect modulatory effect rather than direct inhibition.34
Docking methodology validation
To validate docking accuracy, re-docking of the crystallized ligand molecule against the GSK-3β receptor was performed, yielding an Root Mean Square Deviation (RMSD) of 1.2 Å, which is within acceptable limits for docking reliability.35
Automated screening
After validating the docking approach, an in-silico virtual screening process was conducted through molecular docking simulations. This involved screening a library of compounds against the target protein to identify potential ligands with favorable binding affinities. The compounds were assessed based on their predicted binding modes and energies, allowing for the prioritization of molecules with the most promising interactions with the target protein. This virtual screening approach facilitated the efficient identification of lead compounds for further experimental validation, potentially accelerating the drug discovery process.
Following molecular docking simulations, lead compounds were chosen based on their lowest binding energies, falling within the range of −3 to −13 kcal/mol. Evaluation of the outcomes prioritized the examination of hydrophilic and lipophilic interactions among binding residues within the active ligand binding site of the protein and the ligand. The determination of the ligand’s binding affinity for a particular target was executed utilizing the equation:
Ki = e[ΔG/RT]. Here, ΔG represents the alteration in free energy upon binding, R denotes the gas constant, and T signifies the temperature in Kelvin.
Results
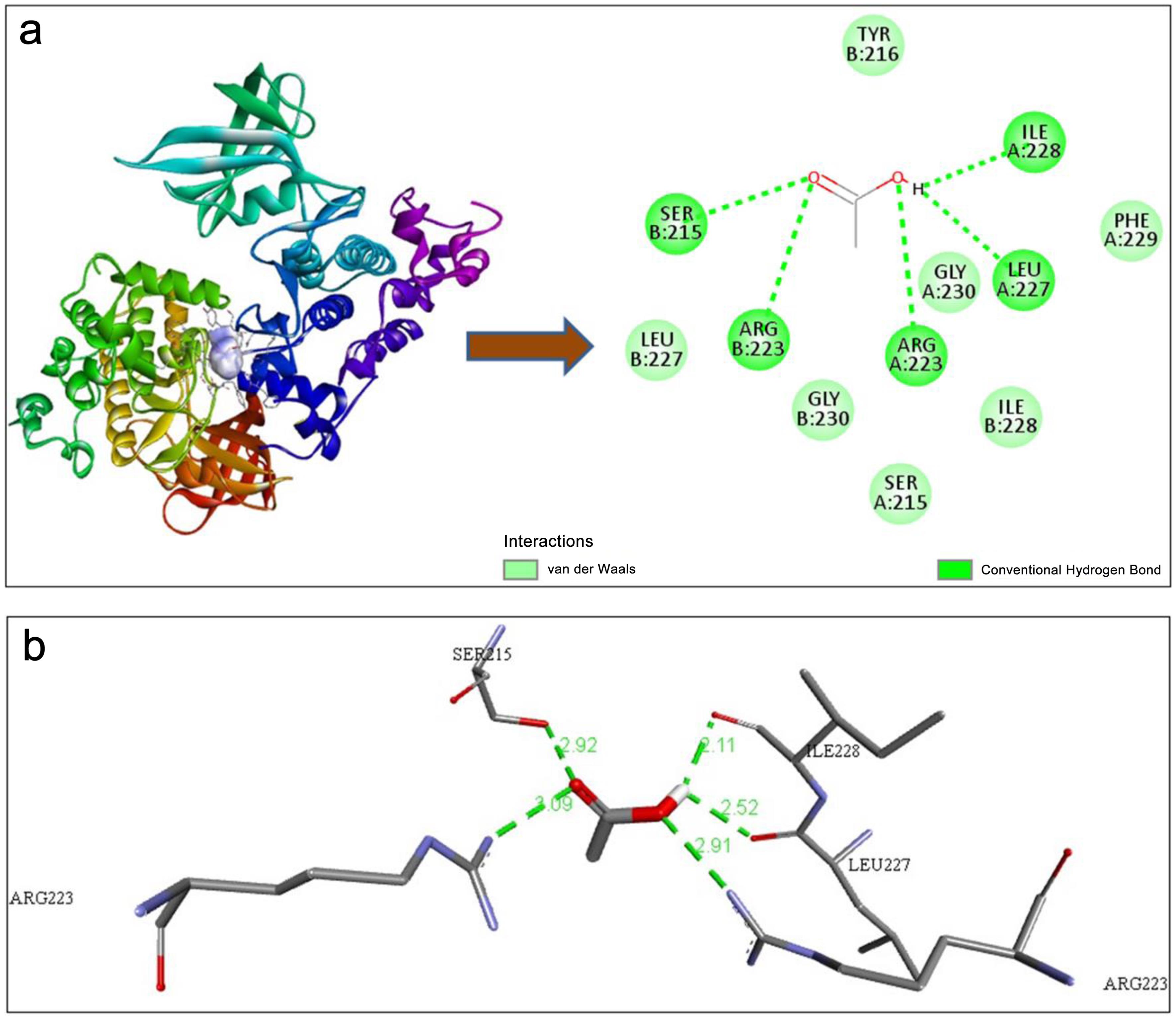
Figure 1c shows nine modes of interaction of the ligand with the protein. Cadmium acetate is able to bind with the enzyme GSK-3β in nine modes. The mode 1 interaction of Cd creates two different bonds: a conventional hydrogen bond and Van der Waals interactions with various amino acids of chain A and chain B of the enzyme GSK-3β, as shown in Figure 2a. The mode 1 interaction of Cd with the enzyme GSK-3β forms five conventional hydrogen bonds, as shown in Figure 2b. The bonds are formed with Ile A:228, Leu A:227, Arg A:223, Arg B:223, and Ser B:215 of the enzyme, and their respective bond distances are 2.11 Å, 2.52 Å, 2.91 Å, 3.09 Å, and 2.29 Å, respectively.
The “Mode of Interaction” describes how GSK-3β and Cd are interacting. Here, it’s denoted as “1,” likely a categorization within a specific interaction scheme. This implies a well-defined and characterized interaction mode based on computational modeling. It was found that Ile A:228, Leu A:227, Arg A:223, Arg B:223, Ser B:215, Phe A:229, Gly A:230, Ile 228, Ser A:215, Gly B:230, Leu B:227, and Tyr B:216 amino acid residues of the enzyme are involved in the formation of conventional hydrogen bonds and Van der Waals interactions. The binding energy of −3.4 kcal/mol mentioned here indicates a favorable interaction, where the formation of the protein-ligand complex releases energy. Quantifying this energy is crucial for comprehending the thermodynamics of the interaction and forecasting its biological significance.
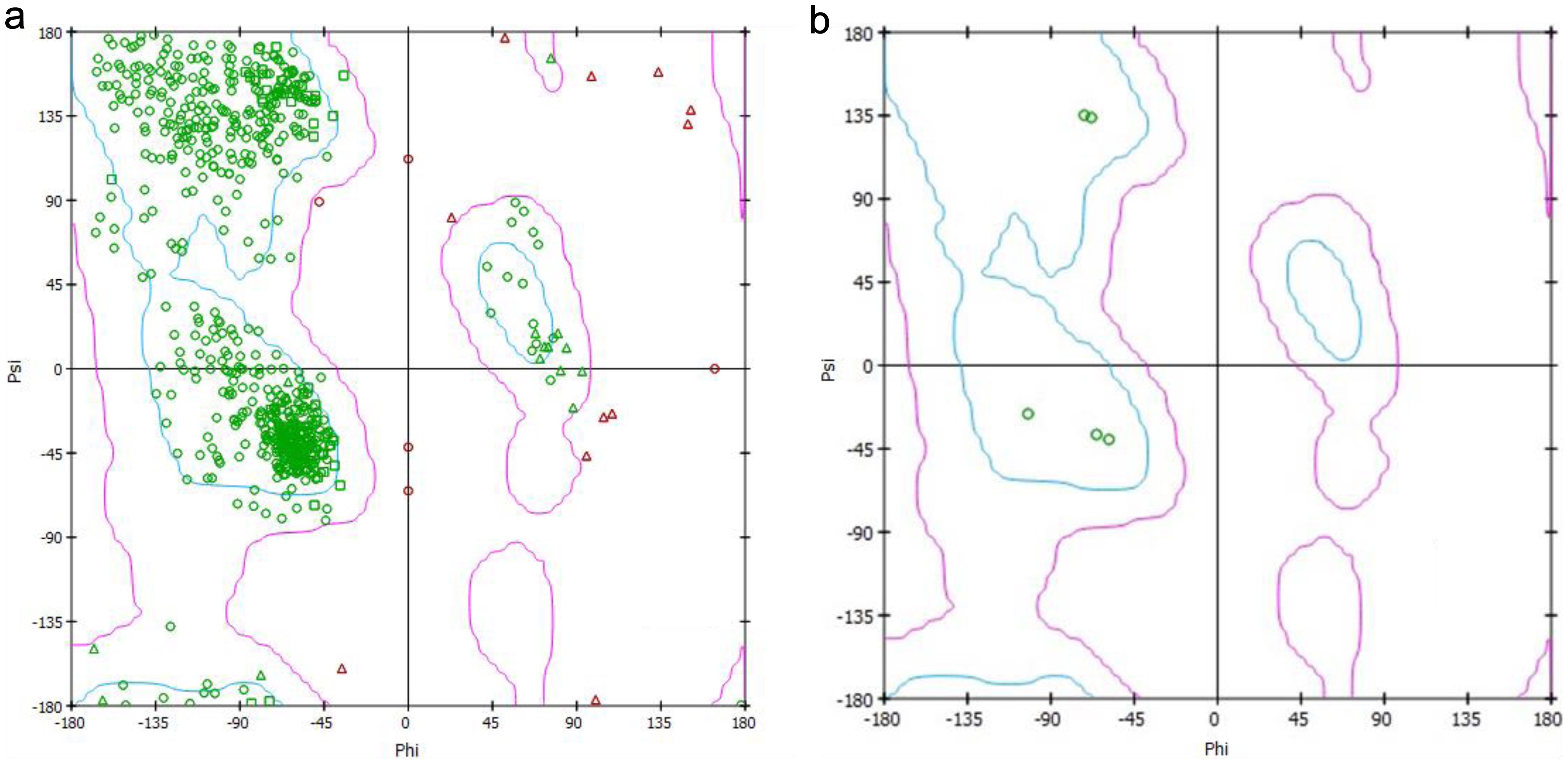
The amino acid distribution of the enzyme is shown in the figure by the Ramachandran plot for the protein in Figure 3a, and the interacting amino acids with the enzyme in Figure 3b. Here, we can see that only five amino acid residues are forming bonds with the ligand.
The observed binding energy of −3.4 kcal/mol in Table 1 indicates a weak interaction. However, this may still hold relevance given the cumulative effects of cadmium exposure over time. Chronic exposure may lead to allosteric modulation or indirect activation of GSK-3β, as suggested by studies linking environmental toxins to enzyme dysregulation.36
Molecular interaction involving the protein glycogen synthase kinase-3 and the ligand cadmium
| Protein | Ligand | Mode of interaction | Interacting residues | Types of bonds | No. of Hydrogen bonds | Binding energy (kcal/mol) |
|---|---|---|---|---|---|---|
| GSK-3β | Cadmium acetate | 1 | Ile A:228, Leu A:227, Arg A:223, Arg B:223, Ser B:215 | Conventional Hydrogen Bond | 5 | −3.4 |
| Phe A:229, Gly A:230, Ile 228, Ser A:215, Gly B:230, Leu B:227, Tyr B:216 | Van der Waals |
Discussion
The results section provides valuable insights into the molecular interaction between the protein GSK-3β and the ligand Cd, shedding light on the specific residues involved, types of bonds formed, and the strength of the interaction. The analysis reveals that Cd is capable of binding with the enzyme GSK-3β in nine distinct modes, as depicted in Figure 3. This highlights the versatility of the ligand in interacting with the protein, suggesting multiple potential binding sites and modes of action. Understanding these diverse interaction modes is crucial for elucidating the complex molecular mechanisms underlying Cd’s effects on GSK-3β activity and associated downstream signaling pathways. The study by Jacobs et al.20 also discussed that GSK-3β plays a pivotal role in modulating diverse signaling pathways governing cellular destiny. Its activity can be augmented by phosphorylation at Tyr-216 by enzymes such as ZAK1 or Fyn. In Figure 2, we have also found that Tyr B:216 has a type of interaction. Upon phosphorylation, GSK-3β becomes more active, exerting its influence on downstream targets.37 Notably, GSK-3β can phosphorylate substrates like β-catenin, marking it for degradation via the ubiquitin-proteasome system. This regulatory mechanism underscores the intricate control exerted by GSK-3β over crucial cellular processes related to the formation of AD.38 Hydrogen bonds are specific interactions that occur between a hydrogen atom covalently bonded to an electronegative atom (e.g., oxygen or nitrogen) and another electronegative atom with lone pair electrons. These bonds play a crucial role in stabilizing ligand-protein complexes by contributing specificity and directional strength.39 Van der Waals interactions, on the other hand, are non-specific, weak forces arising from transient dipoles in atoms and molecules. These interactions help to optimize the fit of the ligand within the protein’s binding pocket, enhancing overall binding stability despite their weaker individual contributions compared to hydrogen bonds.40
Figure 2a, b, and the data presented in Table 1 show the number of hydrogen bonds formed between GSK-3β and Cd. Hydrogen bonds play a crucial role in stabilizing protein-ligand complexes by providing specificity and strength to the interaction. Hydrogen bonds are relatively strong electrostatic attractions between a hydrogen atom and an electronegative atom (like oxygen or nitrogen).41 Van der Waals interactions are weaker, arising from transient dipole moments in molecules. Knowing the types of bonds involved helps elucidate the nature and strength of the interaction.42 The formation of five conventional hydrogen bonds between Cd and GSK-3β, as depicted in Figure 2a and b, underscores the specificity and strength of the interaction. These hydrogen bonds, established with residues such as Ile A:228, Leu A:227, Arg A:223, Arg B:223, and Ser B:215, contribute to the stability of the protein-ligand complex by facilitating directional interactions between the ligand and the protein’s active site. The study by Shahab et al.43 suggests that the precise bond distances reported provide valuable structural insights into the geometry of the interaction, aiding in the rational design of potential inhibitors or modulators targeting this interaction interface.
Figure 3 presents Ramachandran plots illustrating the distribution of total amino acids in GSK-3β (Fig. 3a) and the specific amino acids interacting with Cd in mode 1 (Fig. 3b). This analysis reveals that only five amino acid residues participate in forming bonds with the ligand, emphasizing the selectivity of the interaction and the importance of these residues in mediating the protein-ligand interaction.44 The spatial arrangement of these residues within the protein’s structure, as shown in our study, provides crucial insights into the determinants of binding specificity and affinity.
The quantification of binding energy is essential for assessing the thermodynamic stability of the interaction and predicting its biological relevance.45 The negative value suggests that the binding process releases energy, indicating a spontaneous and potentially high-affinity interaction between the protein and the ligand.46 In our study, the binding energy of −3.4 kcal/mol indicates a favorable interaction between GSK-3β and Cd, where the formation of the protein-ligand complex is energetically favorable. This energy quantification is vital for understanding the thermodynamics of the interaction and predicting its biological relevance.
The library of ligand interaction modes provides critical insights into the potential binding affinities and interaction profiles of ligands with the GSK-3β enzyme. Such information is instrumental in identifying lead compounds for therapeutic interventions. Previous studies have shown that detailed interaction libraries can streamline the drug development process by highlighting ligands with optimal pharmacological profiles.47,48 The selected ligands exhibited favorable physicochemical properties, supporting their potential as drug candidates for Alzheimer’s therapy. Studies have emphasized the importance of draggability predictions in streamlining early-stage drug discovery.49
Wang et al.50 introduced a significant innovation in their method by incorporating an atomic binding score. This score enables researchers to systematically inspect and enhance compounds in a structure-based drug design framework. Reynolds and Reynolds demonstrated the feasibility of establishing a group-equivalent scheme for ligand binding, albeit within the constraints of closely related proteins,51 particularly in terms of size. This discovery carries significant implications for drug design, encompassing both experimental and computational perspectives. Moreover, it paves the way for developing a more comprehensive approach to evaluate ligand binding efficiency. GSK-3β’s catalytic activity undergoes regulation through phosphorylation at two distinct sites: Ser 9 and Tyr 216. Phosphorylation of Ser 9 deactivates GSK-3β, whereas phosphorylation at Tyr 216 within the activation loop enhances its catalytic activity. In this study (Fig. 2a and Table 1), we found a Van der Waals bond in the B chain of the GSK-3β enzyme with Tyr 216 of the ligand, which may be a potential area for new drug discovery. Further, we found that as Cd forms five conventional hydrogen bonds in Figure 2a, b, and Table 1, designing a potential drug will be more effective if a drug is selected based on a higher number of hydrogen bond-forming agents with the GSK-3β enzyme than cadmium to prevent AD.
The interaction of cadmium acetate with key residues such as Arg141 and Lys183 may disrupt GSK-3β’s regulatory mechanisms. Computational studies suggest that such interactions can lead to conformational changes affecting enzyme activity.52 Further experimental work will be needed to confirm these mechanistic effects.
Limitations and future directions
While molecular docking provided valuable insights, incorporating molecular dynamics simulations could further elucidate the binding stability and conformational dynamics of the ligand-GSK-3β complex. Future studies will incorporate molecular dynamics simulations to enhance the understanding of these interactions.53 Additionally, while this study highlights the cadmium acetate-GSK-3β interaction through in silico methods, validation through in vitro and in vivo studies is essential to establish the biological significance of these findings. Future work will focus on experimental approaches such as isothermal titration calorimetry, surface plasmon resonance, or kinase activity assays to confirm these interactions.54 The in silico methods employed provide valuable insights into the molecular interactions between cadmium acetate and GSK-3β, emphasizing the need to understand the molecular mechanisms underlying pollutant-induced neurotoxicity. Further exploration into stronger drug candidates with enhanced binding interactions with GSK-3β, surpassing those of cadmium acetate, holds promise for mitigating the neurological effects of environmental toxins and combating AD and related conditions.
Conclusions
This study provides novel insights into the role of cadmium acetate-induced GSK-3β dysregulation in AD using molecular docking. By identifying key ligand interactions and binding sites, this research lays the groundwork for further exploration into GSK-3β inhibitors as potential therapeutic agents. The urgency of addressing Alzheimer’s disease underscores the importance of identifying novel targets and ligands to accelerate drug development. The findings elucidate the intricate relationship between environmental pollutants, neurological disorders, and the dysregulation of GSK-3β in AD pathogenesis. Cadmium acetate is highlighted as a significant environmental toxin capable of inducing GSK-3β dysregulation through various mechanisms, thereby contributing to the neurodegeneration observed in AD. The in-silico methods employed provide valuable insights into the molecular interactions between cadmium acetate and GSK-3β, emphasizing the need to understand the molecular mechanisms underlying pollutant-induced neurotoxicity. Further exploration into stronger drug candidates with enhanced binding interactions with GSK-3β, surpassing those of cadmium acetate, holds promise for mitigating the neurological effects of environmental toxins and combating AD and related conditions.
Declarations
Acknowledgement
The authors express their gratitude to the Department of Human Physiology at Tripura University (A Central University of India) for providing the necessary instruments that facilitated the smooth conduction of this study.
Ethical statement
The study was conducted in accordance with the principles outlined in the Declaration of Helsinki. This computational study involved no human or animal subjects, requiring no IRB or animal care approvals, and adheres to ethical guidelines for computational research. The individual consent was waived.
Data sharing statement
All the data are formed during our experiment are not submitted anywhere and will be shared on request.
Funding
Not applicable.
Conflict of interest
The authors declare that they have no competing interests.
Authors’ contributions
Experiment assistance (SP), analysis (SP), data visualization (SP), draft of the manuscript (SP, SB), experiment design (SB), and experiment performance (SB). All authors have reviewed and approved the final version of the manuscript.

 Author information
Author information
![(a) 3D structure of glycogen synthase kinase-3 (GSK-3β) enzyme (PDB ID 1i09); (b) Structure of Cadmium acetate [PubChem compound identifier 10986]; and (c) Areas of interaction of Cadmium with the enzyme glycogen synthase kinase-3. (a) 3D structure of glycogen synthase kinase-3 (GSK-3β) enzyme (PDB ID 1i09); (b) Structure of Cadmium acetate [PubChem compound identifier 10986]; and (c) Areas of interaction of Cadmium with the enzyme glycogen synthase kinase-3.](https://publinestorage.blob.core.windows.net/ea0075ea-a06a-4b6a-b4c2-ee5a089a76ad/ncs-2024-00045-g001.jpg)