Introduction
COVID-19 has spread in many countries through human-to-human transmission and rapidly escalated into a global crisis within the initial few months.1 Severe COVID-19 patients require admission to the intensive care unit (ICU), oxygen, mechanical ventilation, and reach without getting any urgent medical care.2 As of September 20, 2023, the World Health Organization (WHO) has recorded a total of 761,769,759 confirmed cases of COVID-19, with 6,784,181 reported fatalities worldwide.3 The transmission dynamics of COVID-19 are shaped by a combination of environmental, demographic, social, and biological factors. Environmental aspects like climate, temperature, humidity, and air pollution affect viral stability and spread, with cold, dry conditions enhancing transmission. Dense populations and urbanization facilitate the virus’s rapid diffusion due to close human contact, while high mobility and migration further exacerbate spread across regions. Social behaviors, such as gatherings in close contact settings, and economic disparities contribute to unequal risks, especially in communities with limited access to healthcare and crowded living conditions. Biological factors include asymptomatic transmission, viral mutation, pre-existing health conditions, and increased vulnerability, while public health interventions like lockdowns and vaccination efforts are crucial in controlling outbreaks. However, gaps in healthcare access and vaccination disparities can prolong the pandemic and heighten the risk of new variants emerging. These dynamics create a complex system that influences how novel coronaviruses diffuse within societies.4,5 However, this pandemic had an unprecedented impact on global health, economies, and daily life. In response to this monumental crisis, the scientific community has swiftly mobilized to develop, refine, and deploy diagnostic tools for virus detection. The pandemic has shown the critical role of diagnostics in the timely identification, containment, and management of infectious diseases. However, the global healthcare landscape has been profoundly affected by the strain placed on resources, logistical hurdles, and shifting priorities brought about by the pandemic. As a result, diagnostic activities have faced unprecedented challenges, ranging from supply chain disruptions to workforce shortages, reducing testing capacities, and patient care delays.6,7
As the pandemic has evolved, new challenges have emerged, particularly the appearance of variants of concern (VOCs), highlighting the need for adaptable and effective diagnostic strategies.8,9 Keeping pace with the latest advancements in COVID-19 diagnostics is crucial. This study presents an in-depth literature review of existing molecular and immunoassay-based diagnostic techniques, elucidating their strengths, limitations, and the emergency regulatory approvals they have received. Additionally, it briefly outlines novel diagnostic methods that may prove valuable in future pandemics. This review is indispensable for staying informed about current diagnostics for COVID-19, enhancing preparedness for future pandemics, and strengthening our collective resilience against global health threats.10–13
Study design or methods
We searched PubMed (www.ncbi.nlm.nih.gov/pubmed ) for full-text articles by using the keywords “COVID-19”, “SARS-CoV-2”, “Diagnostics,” “Variants of concern,” “Immunoassay -based diagnosis techniques,” “Imaging-based diagnosis,” and “Molecular assay-based diagnosis techniques.” Then, the collective literature was examined and presented in this narrative review. Additionally, the data were obtained from analyzing publicly available datasets http://www.io.nihr.ac.uk/report/covid-19-diagnostics/ , https://gisaid.org/ , and https://ourworldindata.org/coronavirus . The software Biorender is used to draw some elements (https://www.biorender.com/ ) of a few figures.
COVID-19 pandemic current global scenario
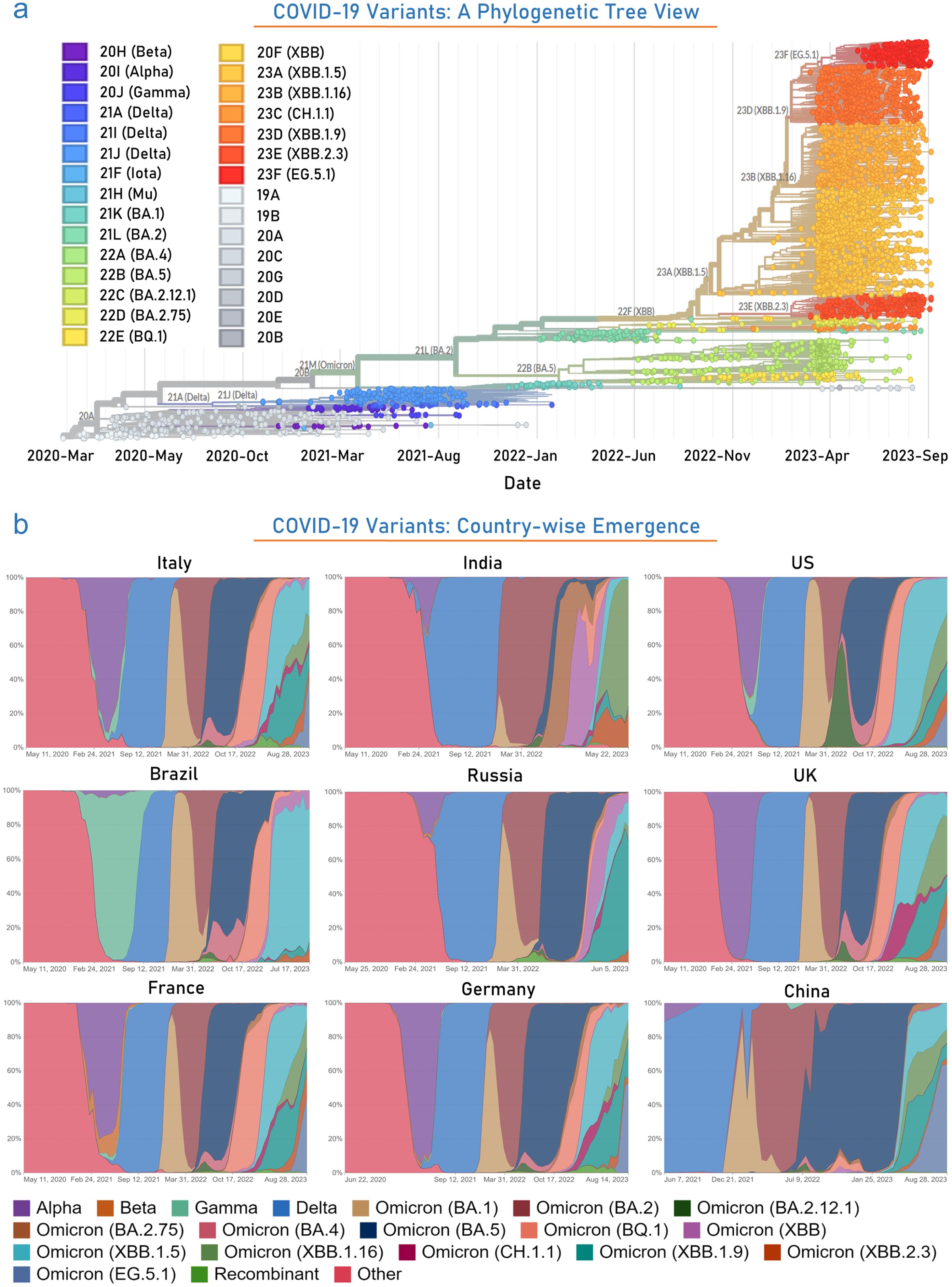
The earliest coronaviruses were studied in human patients with the common cold, initially identified as human coronavirus OC43 and human coronavirus 229E.14 Subsequently, other human coronaviruses were discovered, including SARS-CoV (2003), HCoV NL63 (2004), HKU1 (2005), MERS-CoV (2012), and SARS-CoV-2 (2019). Most of these viruses are associated with severe respiratory infections, and the deadliest variants have caused MERS, SARS, and the COVID-19 pandemic.15 The progression of this pandemic has typically followed a pattern of waves characterized by sudden surges in new cases followed by declines. This pattern may result from various factors, including infection prevention measures, host activities, the time-dependent efficacy of vaccines, and viral mutations. As an adaptive mechanism, coronaviruses frequently mutate, often resulting in variants with altered properties that can lead to higher infection rates and increased disease severity. Furthermore, these variants may evade detection and develop resistance to vaccines. Based on associated risk factors, the WHO has categorized these into VOCs: alpha, beta, gamma, delta, and omicron, and Variants of Interest (VOIs): Lambda and Mu (Fig. 1a). Alpha/B.1.1.7 (+S:484K and +S:452R mutations, respectively) was first reported in September 2020 in the United Kingdom and was estimated to have 40 to 80% higher transmissibility than the wild-type strain. Beta/B.1.351 variant mutated at +S:L18F was documented in May 2020 in South Africa. The gamma/P.1 variant detected in Brazil in November 2020 has 17 amino acid replacements (N501Y, K417T, and E484K are of concern) and is twice extra transmissible and 50% more lethal than the previous strain. The delta/B.1.617.2 variant, first discovered in October 2020 in India, was identified as the most infectious virus with 50% more transmissible power and has significant spike protein mutations, including D614G, T478K, L452R, and P681R. Recently, in November 2021, the Omicron/B.1.1.529 variant was identified in South Africa, and it has higher transmissibility than the other variants. It has 60 mutations, including 8 synonymous, 50 non-synonymous, and 2 non-coding mutations.16–19 The predominant emergence of the Omicron variant started from mid-December 2021 onwards. Till now, several variants of omicron have been reported. Recently, a study reported the presence of omicron in 99.5% of sequenced samples in the US during this short duration (December 2020–January 2022).20 BA.2.86, a variant of SARS-CoV-2 with around 30 mutations enhancing immune evasion, did not dominate in late summer/fall 2023. Its descendant, JN.1, has emerged with increased transmissibility and immune evasion. JN.1 cases coincide with a rise in overall COVID-19 cases. Symptoms are similar to previous omicron variants, with anecdotal reports of more diarrhea. The infectious period mirrors other omicron variants. Older vaccines offer limited protection due to genetic differences and waning immunity, requiring adjustments akin to annual flu vaccines to combat evolving variants effectively.21,22
(a) Figure illustrates the several emerged and developed VOCs in the phylogenetic tree view, spanning from March 2020 to September 2023. (b) The figure represents the extent and duration of infection due to several variants in the top-most countries throughout this COVID-19 pandemic. Data were obtained from the publicly available datasets https://ourworldindata.org/coronavirus , and https://gisaid.org/ .
To determine the disease severity/mortality in the US, the CDC has analyzed the data from three different COVID-19 pandemic periods, i.e., (i) December-2020 to February-2021 (winter of 2020–2021), (ii) July to October-2021 (Delta) and (iii) December-2021 to September-2023 (Omicron with its mutations). This study determines the severity and mortality of the disease by comparing the daily reported cases, emergency department (ED) visits, hospital admissions, and occurrence of several deaths during Omicron vs. Delta periods of COVID-19. The changes observed during Omicron compared to winter to the delta in the daily number of cases, ED visits, hospital admissions, and deaths were 219%, 137%, 31%, and −46%, respectively, compared to the delta period. These variations differed by 386%, 86%, 76%, and −4%, respectively. CDC has also observed many changes in emergency visits and hospital admissions of children and adolescents during omicron prevalence. Furthermore, the occupancy of hospital inpatient beds in the omicron period was 3.4 and 7.2% higher than in the winter 2020–2021 and Delta period, respectively. The occupancy of ICU beds in the omicron period was 0.5% less than the winter 2020–2021 period and 1.2% higher than the Delta period. Based on ICU and hospital inpatient admissions, it concludes that the omicron variant has higher disease severity than its previous variants. However, the unvaccinated individuals and pre-infected individuals were documented to have a higher risk from this omicron variant. Hence, proper vaccination and early diagnosis are the only ways to mitigate the severity and causalities due to this lethal infection (Fig. 1b).23,24
SARS-CoV-2 continues to spread globally, creating a dire situation as it impacts human populations in waves, leading to fluctuating numbers of cases and deaths.25,26 We have analyzed data collected from March 2020 to September 2023 from the top five affected countries, categorized by the total number of confirmed cases. The United States of America, India, France, Germany, and Brazil have reported approximately 108 million, 44 million, 40 million, 38 million, and 37 million confirmed cases, respectively. These countries have recorded 1.2 million, 0.5 million, 0.15 million, 0.175 million, and 0.7 million deaths, respectively. Most nations have experienced the first and second waves of infections, with some facing a third wave. It has been observed that the cumulative number of confirmed cases and deaths per day during the second and third waves has increased exponentially compared to the previous wave.3 Protecting people from COVID-19 remains a significant challenge due to inadequate diagnostics. High-quality diagnostics are essential to curb the spread and severity of the disease. Understanding the biology of COVID-19 is crucial for accurate diagnosis. This section will elucidate the virus’s interaction with the human body and its significance in developing effective diagnostic strategies.
Current landscape of SARS-CoV-2 biology
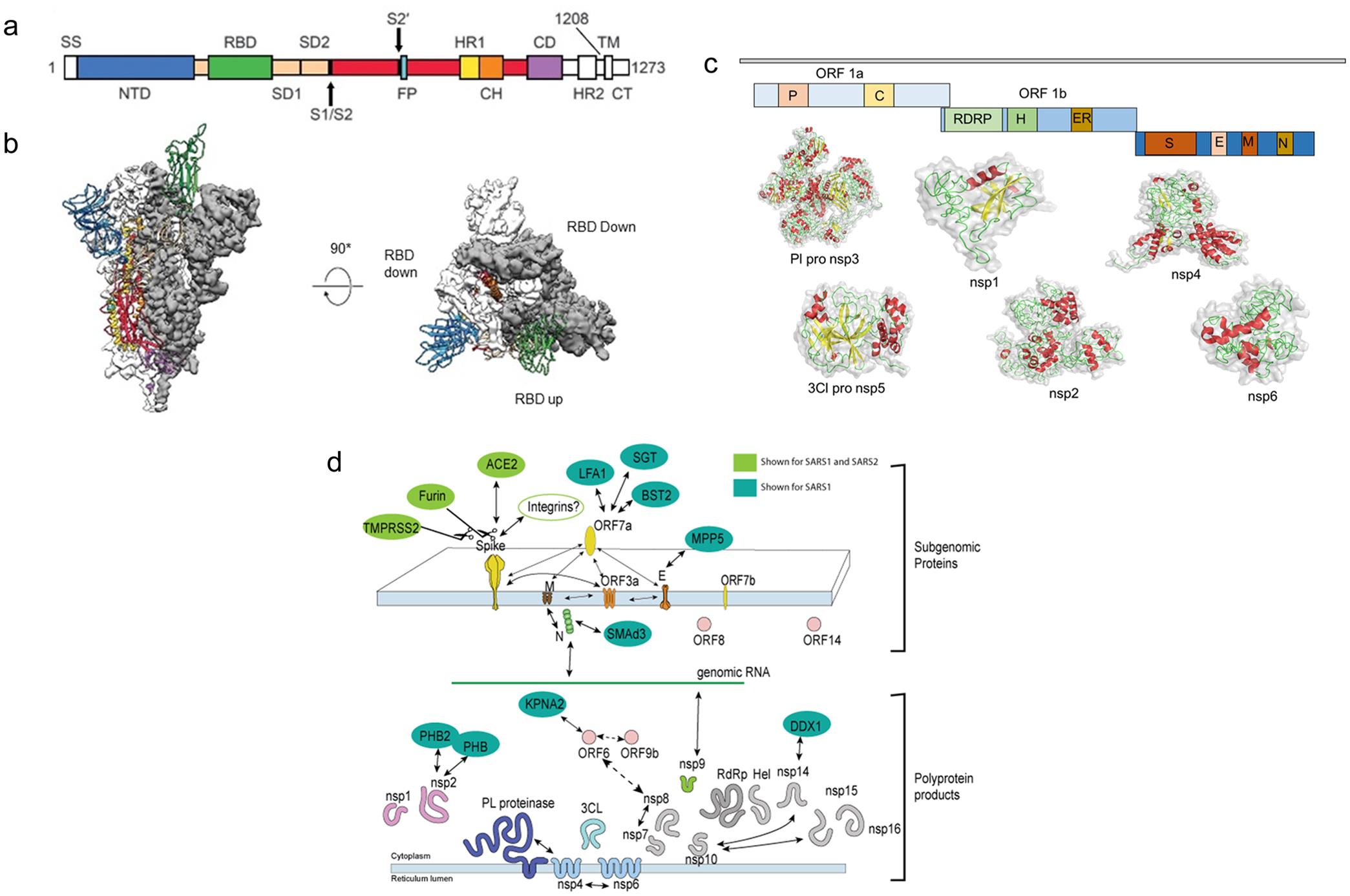
Coronaviruses include many enveloped positive-sense ssRNA viruses with spherical shapes and distinct spiked surface projections. Owing to the average 26 to 32 kb genome size, SARS-CoV-2 has been classified as the largest RNA virus with a diameter range of 60–140 nm. Furthermore, the average size of the envelope and spikes were ∼80 nm and ∼20 nm, respectively.15,27 CoV envelope is made up of a lipid bilayer anchoring structural envelope (E), membrane (M), and spike (S) proteins, which protect the virus from the harsh environment outside the host (Fig. 2).28 Beta-coronavirus subgroup A, a coronavirus variant, also characteristically includes hemagglutinin esterase (HE), a short spiky-surface protein.29 Severe acute respiratory syndrome coronavirus 2, widely known as SARS-CoV-2, has been acknowledged as a beta-coronavirus and has 96% homology with bat CoVs and ∼70% similarity with SARS-CoV.29,30 The major genomic distinction of these enveloped viruses includes the presence of a positive ssRNA genome, helical nucleocapsid, 5′ methylated cap, and 3′ poly (A) tail. The nucleocapsid (N) consists of several N protein copies attached to the RNA in a constant beads-on-a-string configuration. The genomic organization of CoVs is represented as “5′-leader-UTR- transcriptase/replicase-spike-envelope-membrane-nucleocapsid -3′UTR-poly (A) tail”. The development of quality diagnostic tools against SARS-CoV-2 infection entirely depends upon the knowledge of viral biology. In this section, we have delineated the biological role of the various SARS-CoV-2 structural components. ORF1a and ORF1b genes are responsible for coding for transcriptase/replicase polyproteins, which cleaves into all nonstructural (NS) proteins.31
(a) Figure depicts multiple domains of coronavirus; (b) Receptor binding domains (RBD’s); (c) Computational model of various non-structural proteins (NSP’s) viz., nsp1, nsp2, PL-pro nsp3, nsp4, 3CL-pro nsp5, and nsp6; (d) Various sub-genomic proteins and their interaction-based functions and poly-protein products in the biology of SARS-CoV-1 and 2.
Nonstructural proteins (NSPs)
Nsp1 acts as an inhibitor of the endogenous host translation pathway by forming a complex with the host’s 40S ribosomal subunit, which triggers endo-nucleolytic cleavage near the 5′UTR region of host mRNAs, leading to their degradation. A 5′-end leader sequence in the viral mRNA renders it resistant to NSP1-induced endo-nucleolytic cleavage, thereby protecting it from degradation. This effective inhibition of host gene expression by NSP1 aids the virus in evading the host’s immune response.32,33 Nsp2 plays a role in regulating host cell survival signals through its interaction with PHB1 and PHB2, which modulate the functionality of host mitochondria and protect cells from various stress signals.34 PL-PRO, a domain of SARS-CoV NSP3, is a crucial CoV enzyme involved in the expression and N-terminal cleavage of viral replicase polyproteins, facilitating continuous viral spread. It also cleaves post-translational modifications of host proteins to dodge the antiviral immune response. Additionally, PL-PRO possesses deISGylating and deubiquitinating activities and regulates Lys-48 (K48) and Lys-63 (K63) linked polyubiquitination, further contributing to viral evasion mechanisms.35,36 The NSP3 is a large and multifunctional protein encoded by the CoV genome. Along with PL-pro, NSP3 encompasses multiple other domains (viz., macro domain, ubiquitin-like domain, N-terminal acidic domain, middle domain, and C-terminal domain) with their diverse functions. Macro domain is involved in ADP-ribose binding and has been implicated in antagonizing host immune responses.37–39 The ubiquitin-like domain is involved in protein-protein interactions and may play a role in host cell manipulation. The N-terminal acidic domain is implicated in interactions with host cell proteins and may contribute to the modulation of cellular processes. The middle domain contains various motifs and may have roles in protein-protein interactions, RNA binding, and possibly other functions. The C-terminal domain has been suggested to be involved in membrane association and may play a role in viral replication complex formation. These domains, along with the PL-pro domain, collectively contribute to the multi-functionality of NSP3 and are crucial for the virus to effectively replicate and evade host immune responses.40–42
NSP4, in association with NSP3, induces viral replication by aiding the assembly of the viral cytoplasmic double-membrane vesicles. Moreover, NSP4 averts the host cell’s NF-kB signaling and inhibits dimerization, phosphorylation, and nuclear translocation of the host IRF3, antagonizing the type I interferon-induced host innate immune response.43 Nsp5/Proteinase 3 moiety (3CL-Pro) is one of the major cysteine proteases found in CoVs, catalytically cleaving the C-terminus of the viral replicase polyprotein at 11 conserved sites. It recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. It is a member of the MEROPS peptidase C30 family, forming a catalytic dyad with its active histidine and cysteine site residues.37–39 Nsp6 is responsible for early autophagosome induction from the host endoplasmic reticulum. In addition, NSP6 also restricts the expansion of nonfunctional phagosomes that are incompetent in delivering viral particles to lysosomes.44 Eight subunits of NSP7 and NSP8 combine in a hollow cylindrical-like hexa-decamer arrangement to participate in viral replication as a primase and synthesize lengthier products than oligonucleotide primers. NSP8 has conserved D/ExD/E motifs at N and C-terminals, among which the N-terminal motif, being a part of the Mg2-binding active site, is crucial for the RNA polymerase function.45 Nsp9 promotes viral replication by acting as a single-strand RNA-binding protein. The proteins consist of highly conserved N-finger and GXXXG motifs responsible for dimerization. Along with NSP8, it disrupts host immune activity by suppressing cell membrane protein integration.46 Nsp10 aids the viral transcription by regulating the cap methylation of viral mRNAs. It also stimulates the potential functionalities of both 3′-5′ exo-ribonuclease (NSP14) and 2′-O-methyltransferase (NSP16).47 Nsp12/RNA-directed RNA polymerase (RdRp) is produced by OFR1b cleavage and plays a pivotal role in modulating the replication and transcription of the viral genomic RNA. NSP12 polymerase activity is enhanced when it binds with cofactors: NSP7 and NSP8.48 Nsp13/Helicase (Hel) is an Mg-dependent, multifunctional protein having an N-terminal zinc-binding domain that presents nucleic acid duplex- uncoiling activity with 5′ to 3′ polarity.49 Nsp14/proofreading 3′-5′ exoribonuclease/Guanine-N7 methyltransferase (ExoN) is a dual activity enzyme that possesses 3′-5′ proofreading exoribonuclease activity and N7-guanine methyltransferase potential in ssRNA/dsRNA. The proofreading activity lowers the viral sensitivity to RNA mutagens.50 Nsp15/Uridylate-specific endo-ribonuclease (NendoU) is a Mn-dependent and uridylate-specific RNA endoribonuclease, which produces 2′-3′cyclic phosphodiester and 5′-hydroxyl terminal by cleaving the RNA. It inhibits activation of host double-stranded RNA sensors like IFIH1/MDA5, PKR, and OAS by degrading the 5′-poly(U) sequence produced during replication of viral genomic and sub-genomic poly (A) tail, restricting subsequent hybridization of poly(U) with the poly(A) sequence.51,52 Nsp16/2′-O-methyltransferase (2′-O-MT) has specific RNA binding potential and is a methyltransferase that regulates the transfer of methyl group from viral mRNA 2′-O-ribose cap to the 5′-cap arrangement. N7-methyl guanosine plays a crucial role in escaping the host immune response as it is essential in NSP16 binding and viral mRNA cap methylation.47,53,54
Spike (S) glycoproteins
Upon binding with the host receptors, S1 glycoprotein anchors the virion to the host cell membrane, instigating the viral infection. The virus binds to the human ACE2 receptor via the S1 protein and gets internalized into the host endosomes, which changes the conformational structure of the spike glycoproteins.55–57 To target human lung cells, it utilizes human TMPRSS2.55 The cathepsin CTSL-mediated proteolysis uncovers the S2 protein fusion peptide, which initiates membrane fusion inside the endosome. S2 protein acts as a class I viral fusion protein by regulating virion and cellular membrane fusion. S2 protein has three distinct structural phases: the pre-fusion native state, the pre-hairpin intermediate state, and the post-fusion hairpin state. During host cell membrane and viral particle fusion, the heptad repeats and arranges into a hairpin trimer, bringing the fusion peptide closer to the C-terminal ectodomain. The structure subsequently drives the viral particle and host cell membrane fusion.58,59 S2′ glycoprotein, a viral fusion peptide, gets unmasked when the S2 protein cleaves during viral endocytosis.58,59
Structural and functional proteins
Protein E functions as a viroporin that modulates assembly and maintains the morphological structure of the virus. Inside the host cell membrane, the E protein self-assembles to form pentameric lipid-protein pores, allowing ion transport. In addition, it also participates in apoptosis induction. E protein also enhances IL-1β overproduction by activating the host NLRP3 inflammasome.60 Protein M is an essential viral envelope protein that interacts with other viral proteins and aids in virus assembly and morphogenesis.61 Protein N packs (+) ssRNA viral genome inside a helical RNP (ribonucleocapsid) and interacts with the viral genome and M protein, contributing to virion assembly. Moreover, it enhances the transcription efficacy of viral genomic and sub-genomic RNA.62,63 Instead of this, N-NSP3 interaction plays a crucial role in SARS-CoV-2 viral genome replication. This interaction is vital for facilitating efficient viral genome replication within infected cells. Understanding the N-NSP3 interaction is essential for unraveling the molecular mechanisms underlying viral replication and could potentially lead to the development of targeted antiviral strategies against COVID-19.64,65
ORF3a plays a role in releasing virion particles by forming viroporin, potassium-sensitive, homo-tetrameric ion channels. Additionally, it boosts the expression of fibrinogen subunits (viz., FGA, FGG, and FGB) in the epithelial cells of the host lung, leading to cell apoptosis. It also reduces the level of IFN-I by phosphorylating the serine residue in the degradation sequence of IFNAR1 (IFN alpha-receptor subunit 1), thereby enhancing its ubiquitination.66–68 ORF6 binds to karyopherin alpha 2 and beta 1 on the host cell membrane, disrupting the formation of the nuclear import complex. This disruption causes the accumulation of import factors in the Golgi/ER membrane, resulting in the loss of nuclear transport, which restricts STAT1 nuclear translocation—a key component of interferon signaling—thereby inhibiting antiviral activity and the expression of interferon-stimulated genes (ISGs).69,70 SARS-CoV-2 infected cells express and store ORF7a intracellularly within the Golgi network, playing a crucial role in the virus’s replication. The biological activities of ORF7a include caspase-dependent apoptosis, p38 MAPK activation, inhibition of host protein translation, and suppression of cell growth, highlighting its significant role in virus-host interactions.71 ORF8, a rapidly evolving protein in SARS-related coronaviruses, is crucial for counteracting the host immune response and increasing transmission rates. It resembles the NS8 gene of bat coronaviruses, known for its critical role in host-virus interactions, yet distinctly different from the SARS NS8a and NS8b genes.72–74 ORF10 region is associated with the beta-coronaviruses but apparently does not have any homologous proteins and suggestively may not have functional protein-coding properties. It may act as an RNA precursor and alternatively regulate other cellular pathways.75,76
The expression level of the ACE2 receptor is comparatively high in tongue epithelial cells, making the oral cavity a potential SARS-CoV-2 site of infection. The surface spike proteins of coronavirus promote their access into the host cells, consequently making spike proteins the major targets of monoclonal antibodies and other modern therapeutic strategies.55 A recent report states that the structural information of SARS-CoV-2 S (spike) protein’s ectodomain trimer obtained using a cryo-EM-based study provided significant information required for the development of diagnostic tools against COVID-19.77,78 Based on this knowledge, diagnostic tools against SARS-CoV-2 infection are designed and used for their detection.79 The list of SARS CoV-2 genes, length, and translated proteins are summarized in Table 1.80,81 The biology of COVID-19 is the foundation upon which diagnostic tools are built. An in-depth understanding of the virus’s biology is essential for developing, validating, and improving diagnostic tests to meet the evolving challenges of SARS-CoV-2 and its variants. In the next section, the authors are willing to briefly outline the various diagnostic methods, their development, present status, and regulatory approval.
List of SARS CoV-2 genes, length, translated proteins, and based diagnostic kit examples
| Gene80,81 | Length | No. of nucleotide | Translated protein | Amino acid length | Developed diagnostic kit |
|---|---|---|---|---|---|
| 5′ UTR | 1–265 | 265 | Non-coding region | – | – |
| ORF1ab | 266–21,555 | 21290 | pp1ab/pp1a | 7,096/4,405 | VIASURE |
| S | 21,563–25,384 | 3822 | S | 1,273 | Sampinute COVID-19 |
| ORF3a | 25,393–26,220 | 828 | ORF3a | 275 | – |
| E | 26,245–26,472 | 228 | E | 75 | Mylab CoviSelf |
| M | 26,523–27,191 | 669 | M | 222 | – |
| ORF6 | 27,202–27,387 | 186 | ORF6 | 61 | – |
| ORF7a | 27,394–27,759 | 366 | ORF7a | 121 | – |
| ORF7b | 27,756–27,887 | 132 | ORF7b | 43 | – |
| ORF8 | 27,894–28,259 | 366 | ORF8 | 121 | – |
| N | 28,274–29,533 | 1260 | N | 419 | Clip COVID, Ellume COVID-19 etc. |
| ORF10 | 29,558–29,674 | 117 | ORF10 | 38 | – |
| 3′ UTR | 29,675–29,903 | 229 | Non-coding region | – | – |
SARS-CoV-2: Current diagnostic approaches, development, and regulatory approval
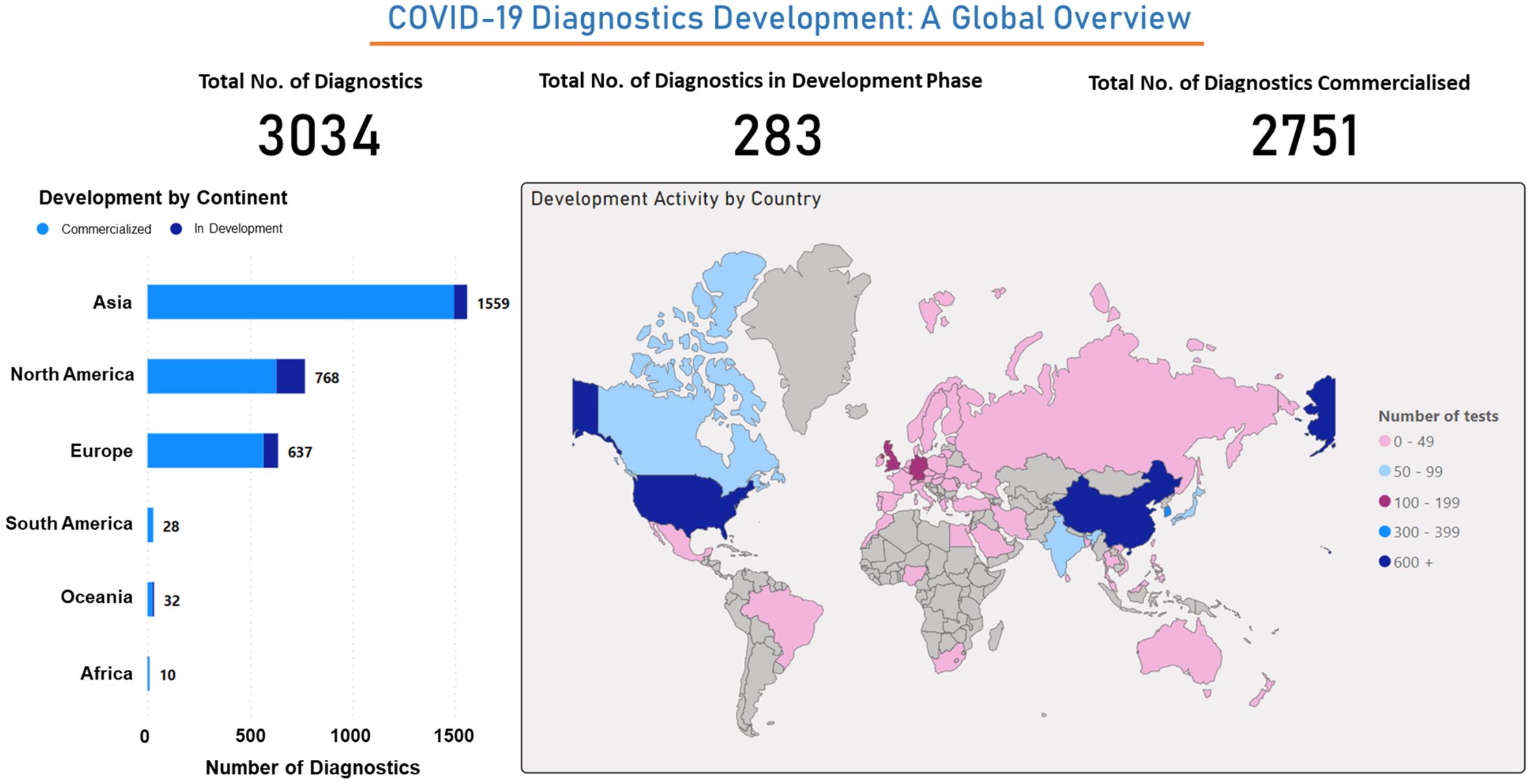
The global In Vitro Diagnostics (IVDs) market for infectious diseases is experiencing substantial growth, primarily fueled by the rising prevalence of infectious diseases. Events such as the COVID-19 pandemic, along with other highly contagious infections like SARS and Ebola virus disease, have spurred rapid advancements in diagnostic technologies as critical components of response strategies. As a result, the global implementation of testing protocols has become a significant driving force in reshaping the diagnostic landscape. Based on data retrieved from a dataset (source), our analysis indicates that out of 3,034 developed diagnostics, 283 are still under development, and 2,751 are commercially available. We have classified these diagnostics by the country’s role in their development, revealing that China and the US are at the forefront. Additionally, Asia emerges as the primary origin of diagnostic development when categorized by continent. In conclusion, the advancement of COVID-19 diagnostics represents a significant achievement driven by the collective efforts of various countries and continents. To address medical challenges related to SARS-CoV-2, the development of rapid diagnostic methods is paramount. Prominent diagnostic techniques include nucleic acid amplification testing (NAAT) for quantifying targeted viral genomic antigens, immunological assays to detect antigenic proteins or immunoglobulins, and biomedical imaging techniques for visualizing disease-related anatomical changes.82 A brief description of these techniques has been given in the following sections:
Molecular diagnostics or NAAT
NAAT is the most sensitive approach for SARS-CoV-2 RNA detection. The chief principle of this method is to amplify specific viral genome regions like spike, envelope, nucleocapsid, genes, and different sections of the first ORF, such as the RdRp gene. Some standard NAAT-based techniques utilized for SARS-CoV-2 diagnosis include RT-PCR, RT-LAMP, NGS, and CRISPR-based assays.82 The list of US-FDA-approved commercially available NAAT kits is summarized in Table 2.
List of US-FDA-approved commercially available nucleic acid amplification test (NAAT) kits
| Kit name | Kit #Cat. No. | Test/kit | Platform | Developer | Detection | Limit of detection (LOD) | Approval | |
|---|---|---|---|---|---|---|---|---|
| Extraction equipment | Amplification equipment | |||||||
| 1COPY COVID-19 QPCR | 444213 | 100 | QIAamp | Light Cycler 480 (Roche) | 1DROP INC. | E, RdRp | 200 | US-FDA, EUA |
| TRUPCR SARS-CoV-2 | 3B304 | 100 | TRUPCR | Rotor-Gene Q 5plex HRM | 3B Blackbio Biotech | RdRp, N, E | 10,000 | US-FDA, EUA |
| 3DMed 2019-nCoV RT-qPCR | 3103010011 | 100 | ANDiS | 7500 RT-PCR | 3D Biomedicine Sci. & Tech. | N, E, ORF-1ab | – | WHO |
| ID NOW COVID-19 | 190-000 | 96 | ID NOW Instrument | Abbott Diagnostic | RdRp | 1,250 | US-FDA, EUA | |
| 09N78-095 | 96 | Alinity m System | Abbott Molecular | RdRp, N | US-FDA, EUA | |||
| Abbott Real Time SARS-CoV-2 | 09N77-090 | 96 | Abbott m2000 | 100 | WHO | |||
| 09N77-095 | 96 | US-FDA, EUA | ||||||
| MassARRAY® | 13279F 13278D 13281D | 96 3840 768 | NucliSENS® easyMAG® | MassARRAY | Agena Bioscience | N, ORF-1. ORF-1ab | 310 | US-FDA, EUA |
| RealStar® | 821025 | 384 | AltoStar® Automation System AM16 | CFX96™ Touch RT PCR | Altona Diagnostic | -ALGenomics | 0.1PFU/mL | US-FDA, EUA |
| BioCode® | 64-C0304 | 384 | NucliSENS® easyMAG® | BioCode® MDx-3000 | Applied BioCode | N | – | US-FDA, EUA |
| Linea™ | DX-1001-001-000 | 100 | QIAamp | QuantStudio™ Dx RT-PCR | Applied DNA Sci. | S | 1,200l | US-FDA, EUA |
| DX-1001-002-000 | 500 | TRIzol™ RNA | QuantStudio 5 RT-PCR | |||||
| DX-1001-003-000 | 1000 | Omega Bio-Tek | 7500 RT-PCR | |||||
| iAMP® COVID-19 | iAMP-COVID19 | 100 | Not required | CFX96 RT-PCR | Atila Biosystem | N, ORF-1ab | 4,000 | US-FDA, EUA |
| BD SARS-CoV-2 Reagents | 445003-01 | 24 | BD MAX™ System | Becton, Dickinson & company | N | – | US-FDA, EUA | |
| Fluorescent RT-PCR | MFG030010 | 50 | TIANamp | 7500/7500 Fast RT-PCR | BGI Europe A/S | ORF-1ab | 150 | WHO |
| MFG030010 | 50 | QIAamp | BGI Genomics | US-FDA, EUA | ||||
| RT-PCR Kit | CT8233 | 48 | Beijing Applied Bio. Tech. | ORF-1ab, N, E | 550 | WHO | ||
| Wantai SARS-CoV-2 RT-PCR | WS-1248 | 48 | Beijing Wantai Bio. Phar. | ORF-1ab, N | 50 | WHO | ||
| BioCore | BC-01-0099 BC-01-0099 x4 | 100 400 | BioCore | N, RdRp | 500 | US-FDA, EUA | ||
| Bio-Speedy | BS-SY-SC2-100 BS-SY-SC2-1000 | 100 1000 | LightCycler 96 | Bioeksen R&D Technologies | ORF-1ab | – | US-FDA, EUA | |
| BioFire | 423745 | 6 | FilmArray® 2.0 | BioFire Defense | ORF-1ab, ORF-8 | – | US-FDA, EUA | |
| 423744 | 30 | |||||||
| BioGX Xfree | 500-003-XMP | 104 | QuantStudio 5 | BioGX | N | 330 | US-FDA, EUA | |
| Biomeme | 3000555 | – | Biomeme’s Franklin RT-PCR | Biomeme | ORF-1ab, S | 1,800GE/mL | US-FDA | |
| Bio-Rad SARS-CoV-2 ddPCR | 12013743 | 200 | MagMAX™ | QX200™ PCR | Bio-Rad | P, N | 630 | US-FDA, EUA |
| Real-Q 2019-nCoV | BS7nCoV | 100 | MagNA Pure 96 | 7500 RT-PCR | BioSewoom | E, RdRp | 6,250 | US-FDA, EUA |
| COVID-19 RT-PCR PNA | TD1100 | 24 | RNeasy Mini kit | BioTNS | N, RdRp | – | US-FDA, EUA | |
| Xpert® | XPRSARS-COV2-10 | 10 | GeneXpert Xpress System | Cepheid | N, E | 250 | US-FDA, EUA | |
| COVID-19 RT-PCR | HBRT-COVID-19 | 24 | KingFisher™ Flex | 7500 RT-PCR | Chaozhou Hybribio Biochem. | N, ORF-1ab | – | WHO |
| Clinomics TrioDx | TR-US-01 | 100 | QIAamp | QuantStudio 6 Flex | Clinomics | RdRp, N, E | – | US-FDA, EUA |
| LOGIX SMART™ | COVID-K-001 | 100 | CoDx Box | Co-Diagnostics | – | 4,290 | US-FDA, EUA | |
| Cue | C1020 | – | Cue Health Monitoring System | Cue Health | N | 20 | US-FDA, EUA | |
| HDPCR™ | 99-57003 | 480 | KingFisher™ Flex | 7500 Fast RT-PCR | ChromaCode | 1,000 | US-FDA, EUA | |
| 2019-nCoV | DA0930 | 24 | QIAamp | Roche Light Cycler | Da An Gene | ORF-1ab, N | – | WHO |
| MobileDetect-BIO BCC19 | MOL4150 | 24 | MD-Bio BCC19 Heater | DetectaChem | N, E | 75,000 | US-FDA, EUA | |
| QuantiVirus | DC-11-0007 | 24 | PureLink™ | Quant Studio 5 RT-PCR | DiaCarta | ORF-1ab, N, E | 100 | US-FDA, EUA |
| DC-11-0017 | 24 | 7500 Fast Dx RT-PCR | ORF-1ab, | US-FDA, EUA | ||||
| Simplexa™ | MOL4150 | 24 | LIAISON® MDX | DiaSorin Molecular | ORF-1ab, S | 242 | US-FDA, EUA, WHO | |
| AMPIPROBE | ENZ-GEN215-0096 | – | GENFLEX platform V1.0 | Enzo Life Sci. | N | 280 | US-FDA, EUA | |
| EURO Real Time | MP 2606-0125 | 25 | QIAamp | LightCycler® 480 II | EUROIMMUN | ORF-1ab, N | 150 | US-FDA, EUA |
| FTD SARS-CoV-2 | 11416302 | 96 | Bio Méreux | 7500 Fast DxReal-Time PCR | Fast Track Diagnostics | – | US-FDA, EUA | |
| 11416300 | 32/96 | 6,250 GE/mL | WHO | |||||
| Advanta | 102-0355 | – | Biomark HD | Fluidigm | N | US-FDA, EUA | ||
| GenePro | CV002 | – | QIAamp | Quant Studio™ | Gencurix | N, E | 5,550 GE/mL | US-FDA, EUA |
| Genetron | RPQ021 RPQ022 | 50 100 | QIAamp DSP | 7500 Fast Dx RT-PCR | Genetron Health | ORF-1ab, N | 1,000 | US-FDA, EUA |
| ePlex® | EA008212 | 12 | GenMark ePlex | GenMark Diagnostics | – | 10 | US-FDA, EUA | |
| COVID-19 RT-Digital | CV0202 | 48 | QIAamp® DSP | QuantStudio™ | Gnomegen LLC | – | 500 | US-FDA, EUA |
| Aptima | PRD-06419 | 250 | Panther System | Hologic | ORF-1ab | 10 | US-FDA, EUA | |
| Hymon™ | 351251 | 96 | QIAamp® DSP | 7500 Dx RT-PCR | HymonBio | N, E | – | US-FDA, EUA |
| Smart Detect™ | COV2-E | 48 | InBios International | N, E, ORF-1ab | 1,100 | US-FDA, EUA | ||
| COVID-19 RT-PCR | JC10223 | 50 25 | Jiangsu Bioperfectus Tech. | ORF-1ab, N | 6,250 | US-FDA, EUA, WHO | ||
| RADI COVID-19 | RV008 | 100 | CFX96 | KH Medical | S, RdRp | – | WHO | |
| KimForest | KF2019CoV01 | 96 | StepOnePlus | KimForest | RdRp | – | US-FDA, EUA | |
| PowerChek™ | R6900TD | – | CFX96 | Kogene Biotech | RdRp, E | 4,000 | US-FDA, EUA | |
| Lucira | 810055970056 | 24 | Disposable Lucira Device | Lucira Health | N | – | US-FDA, EUA | |
| ARIES® | 50-10047 | 24 | Luminex® ARIES® | Luminex | – | 1,000 | US-FDA, EUA | |
| NxTAG® CoV | I054C0463 | 96 | bioMérieux® | Luminex® MAGPIX® | Luminex Molecular Diagnostics | N, E, ORF-1ab | 5,000 | US-FDA, EUA |
| LumiraDx | L018180030096 | – | Qiagen DSP | Roche Light Cycler 480 II | LumiraDx | ORF-1ab | 1,000 | US-FDA, EUA |
| Fluorescent PCR | BUSGN7101109 | 32 | QIAamp | 7500 RT-PCR | Maccura Biotech. | N, E, ORF-1ab | 1,000 | US-FDA, EUA |
| DETECTR BOOST | DETECTRA | 768 | BRAVO BenchCel DB | Mammoth Biosci. | N | 20,000 | US-FDA, EUA | |
| MatMaCorp COVID-19 2SF | ST-CV19-2SF | – | MatMaCorp Solas 8 | DBA Matma | RdRp | – | US-FDA, EUA | |
| Revogene | 410700 | – | REVOGENE SYSTEM | Meridian | N | – | US-FDA, EUA | |
| Accula | COV4100 | – | Accula™ | Mesa Biotech | – | 100 | US-FDA, EUA | |
| MicroGEM Sal6830 | SCF0030 | 30 | MicroGEM Sal6830 | MicroGEM | N, E | – | US-FDA, EUA | |
| DASH | PN-0205 | 768 | DASH Analyzer | Minute Molecular Diagnostics | N | – | US-FDA, EUA | |
| NeuMoDx™ | 300800 | 96 | NeuMoDx™ 288 Molecular | NeuMoDx Molecular | N, Nsp2 | 150 | US-FDA, EUA | |
| Kaira | RDM101-X | 100 | QIA symphony DSP | 7500 Fast RT-PCR | OPTOLANE Tech. | E, RdRp | 2,500 | US-FDA, EUA |
| GeneFinder™ | IFMR-45 | 100 | QIAamp | OSANG Healthcare | E, RdRp, N | – | US-FDA, EUA | |
| OPTI SARS-CoV-2 | 99-57003 99-57004 | – | Duo instrument | OPTI Medical Systems | RdRp, N | 900 | US-FDA, EUA | |
| PerkinElmer® | 2019-nCoV-PCR-AUS | 48 | PerkinElmer® kit | PerkinElmer | N, ORF-1ab | – | US-FDA, EUA | |
| IntelliPlex | 82303-U | 96 | QIAmp | IntelliPlexTM 1000 πCode | PlexBio | E, RdRp, N | 140 | US-FDA, EUA |
| FastPlex | 02.01.1019 | 24 | DropX-2000 | PreciGenome LLC | RdRp, N | 571.4 | US-FDA, EUA | |
| COVID-19 genesis | Z-PATH-COVID-19-CE | 96 | GenoXtract | 7500 Fast RT-PCR | Primer design | ORF-1ab | 330 | WHO |
| Z-COVID-19 | 96 | QIAmp | US-FDA, EUA | |||||
| PhoenixDx | PCCSKU15261 | 50 | Procomcure Biotech | E, RdRp | – | US-FDA, EUA | ||
| PhoenixDx multiplex | PCCSKU15262 | 50 | SphaeraMag | qTower3G | N, ORF-1ab | – | US-FDA, EUA | |
| QIAstat-Dx | 691223 | 6 | QIAstat Dx Analyzer | QIAGEN GmbH | ORF-1ab, RdRp | 500 | US-FDA, EUA | |
| Quest | 39433 | 96 | Roche MagNA Pure-96 | 7500 Fast RT-PCR | Quest Diagnostics | N | 136 | US-FDA, EUA |
| Lyra | CE-M120 | 96 | easyMAG | Quidel | ORF-1ab | 800 | US-FDA, EUA | |
| Solana | M313 | 96 | Solana Instrument | – | US-FDA, EUA | |||
| Rheonix | KCCOV19-24 | 96 | Rheonix Encompass MDx® Workstation | Rheonix | 625 | US-FDA, EUA | ||
| Cobas | 09175431190 | 192 | Cobas 6800/8800 | Roche Diagnostic | ORF-1ab, E | 12 | US-FDA, EUA | |
| 09408592190 | 20 | Cobas Liat | ORF-1ab, N | US-FDA, EUA | ||||
| Nucleic Acid Diagnostic | S3104E | 24 | QIAamp | 7500 Fast RT-PCR | Sansure Biotech. | 200 | US-FDA, EUA | |
| ScienCell™ | RX7038 | 96 | LightCycle | ScienCell | RdRp, N | 3,162 | US-FDA, EUA | |
| STANDARD M nCoV | M-NCOV-01 | 96 | CFX96 | SD Biosensor | ORF-1ab, E | – | US-FDA, EUA | |
| U-TOP™ | SS-9930 | 96 | PANAMAX | – | Seasun Biomaterials | ORF-1ab, N | 1,000 | US-FDA, EUA |
| AQ-TOP COVID-19 | SS-9920 | 96 | QIAamp | CFX96 | 7,000 | US-FDA, EUA | ||
| AQ-TOP COVID-19 PLUS | SS-9940 | 96 | PANAMAX | 1,000 | US-FDA, EUA | |||
| Allplex™ | RP10243X | 100 | QIAamp | 7500 Fast RT-PCR | Seegene | RdRp, N, E | 4,167 | US-FDA, EUA |
| Fosun | PCSYHF03-a | 96 | QIAamp DSP | Shanghai Fosun | ORF-1ab, N, E | 300 | US-FDA, EUA | |
| Fosun 2019-nCoV | PCSYHF | – | – | – | WHO | |||
| Nucleic Acid Detection | GZ-D2RM25 | 50 | QIAamp DSP | 7500 Fast RT-PCR | ORF-1ab, N | – | WHO | |
| SARS-CoV-2 diagnosis | KH-G-M-574-48 | 48 | Nucleic acid extraction | CFX96 | ORF-1ab, N, E | – | WHO | |
| Multiplex RT-PCR | RR-0485-02 | 25 | QIAamp | 7500 Fast RT-PCR | Shanghai ZJ Bio-Tech | – | WHO | |
| Ezplex | GNT2011-1 | 100 | SML GENETREE | RdRp, N | – | US-FDA, EUA | ||
| Talis One | O11200-25 | 25 | Talis One Instrument | Talis Biomed. | ORF-1ab, N | – | US-FDA, EUA | |
| Ex Probe TM | 68020 | – | EZ bead Extraction | TBG Q6000 RT-PCR | TBG Biotech. | RdRp, N, E | 10,000 | US-FDA, EUA |
| SARS-CoV-2 Detection | PGA4102P1/P2 (liquid/lyophilized) | – | – | Tellgen | – | – | WHO | |
| TaqPath | A47813/A47814/A49868 | 200/1000/1000 | MagMAX™ | 7500 Fast RT-PCR | Thermo Fisher | ORF-1ab, N, S | – | US-FDA, EUA |
| TaqPath CE-IVD RT-PCR | A48067 | 1000 | WHO | |||||
| TaqPath pooling | A49918 | 384 | US-FDA, EUA | |||||
| TaqPath RNase P combo | A51333 | 1 | ORF-1ab, N | US-FDA, EUA | ||||
| TaqPath fast PCR combo 2.0 | A51606 | 1 | – | Quant studio 5 flex | US-FDA, EUA | |||
| Amplitude™ TaqPath | A49869 | 20000 | Tecan™ Fluent™ 1080 | Quant studio 7 flex | ORF-1ab, N, S | US-FDA, EUA | ||
| RT-PCR PNA | TD1100 | 100 | RNeasy Mini kit | 7500 Fast RT-PCR | Bio TNS | RdRp, N | US-FDA, EUA | |
| UOL COVID-19 | UOL001 | – | Uh-Oh Labs Point-of-Care Instrument | Uh-Oh Labs | – | – | US-FDA, EUA | |
| ViroKey™ | 300681 | 4050 | – | Sentosa® SA201 | Vela Operations | ORF-1a, RdRp | – | US-FDA, EUA |
| SARS-CoV-2 Test | 801301 | 48 | Xiamen Zeesan | Quant studio 3 RT PCR | Xiamen Zeesan Biotech. | ORF-1ab, N | 200 | US-FDA, EUA |
| Nucleic Acid RT-PCR | SC-COVID19 | 20/100 | MagMAX™ | 7500 Fast RT-PCR | ZhuHai Sinochips Biosci. | 2,000 | US-FDA, EUA | |
| Quick SARS-CoV-2 rRT PCR | R3011 | 1/1K/10K | Bio-Rad CFX96 | Zymo Research | N | 83 | US-FDA, EUA | |
| Clear Dx™ | – | 192 | Hamilton STAR robotic platform, Oxford Nanopore GridION sequencer, and ALPAQUA magnum FLX on deck magnet | Clear Labs | Full Genome | – | US-FDA, EUA | |
| COVIDSeq™ | – | 3072 | NovaSeq 6000 | Illumina | – | US-FDA, EUA | ||
| NextSeq 500 | ||||||||
| NextSeq 550 | ||||||||
| NextSeq 550Dx | ||||||||
| NGS | 102997 | 96 | NextSeq 500 | Twist Biosci. | – | US-FDA, EUA | ||
| NextSeq 550 | ||||||||
| NextSeq 550Dx | ||||||||
This information is obtained from https://www.theglobalfund.org/media/9629/covid19diagnosticproductslist .
RT-PCR
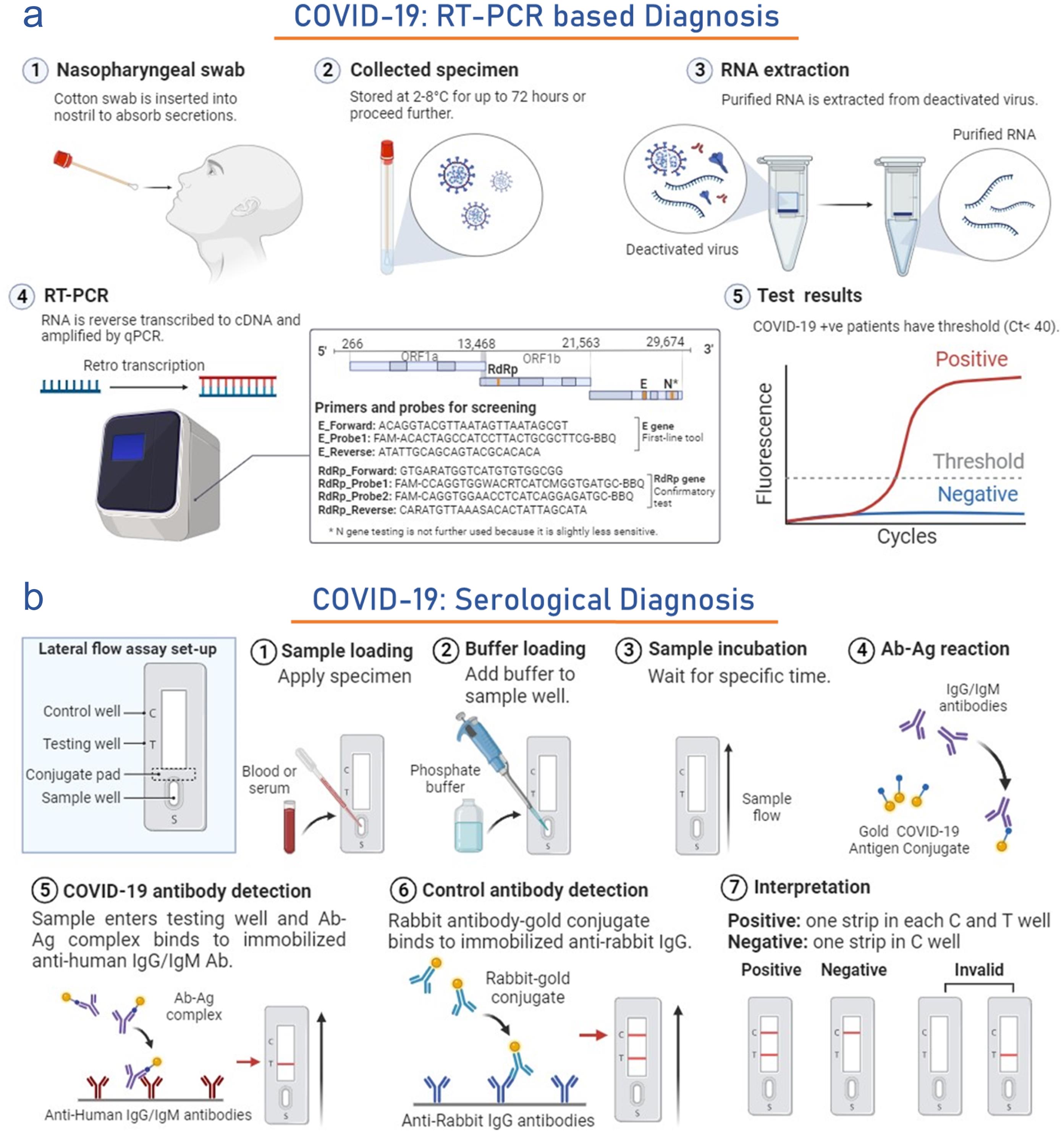
Fast and accurate testing of this infection is considered a significant strategy to control the infection rate in public or hospitals.83 To date, PCR is a major frontline reaction in diagnosing this infection. It requires a set of primers that can be constructed quickly after identifying viral sequences.84 In January 2020, the WHO established and circulated the qRT-PCR protocol to detect this infection. This test is complicated, expensive, and mostly found in large, centralized testing laboratories. Oro-pharyngeal and nasopharyngeal swab tests are two standard methods for specimen collection. Till now, WHO has inaugurated three RT-PCR diagnostic tests targeting genes such as RdRP/Hel, S, and N. The detection of gene E is considered superior and effective to the RdRp gene test.85 Furthermore, a new FDA-approved Abbott ID NOW diagnostic kit has been developed to generate the results within 5 min. The gene detection method of this infection also has limitations and sometimes generates false-negative results; hence, it can be cross-checked by antibody detection. This method is preferable for asymptomatic patients (Fig. 3a).86 Thermo Fisher Scientific (US) created the TaqPathTM COVID-19 Combo Kit, approved by the US FDA for emergency use on March 13, 2020. This kit analyzes nasopharyngeal swabs and bronchoalveolar samples by amplifying S, N, and ORF1ab genes. It can diagnose COVID-19 in 40 min with a 95% detection limit. This means the kit can accurately identify the presence of the virus in samples with a 95% probability, even at low concentrations.87 Similarly, kit which diagnoses nasopharyngeal and throat swabs, Std M nCoV Real-Time Kit (SD Biosensor-Republic of Korea) also approved for emergency use by EUA, US-FDA on 23rd April 2020 targets ORF1ab, RdRp, and Envelop genes at 1–10 copies detection limit and gives result under 30 minutes.88,89
(a) Figure illustrates the process of COVID-19 diagnosis using the real-time RT-PCR. It covers sample collection, RNA extraction, RT-qPCR setup, and result interpretation. This template can be tailored for various RT-qPCR diagnostic protocols. (b) The figure depicts the serologic diagnostic testing of COVID-19, emphasizing the identification of antibodies. It encompasses sample loading, antibody detection, and qualitative test outcomes. The software Biorender (https://www.biorender.com/ ) is used to draw some elements of this figure.
RT-PCR is highly specific and sensitive, establishing it as the gold standard for COVID-19 diagnosis. Detection rates vary by sample type: 63% in nasopharyngeal swabs, 72% in sputum, and 93% in bronchoalveolar lavage fluid.90 However, several challenges accompany RT-PCR-based diagnosis, including the generation of false positive and negative results, high diagnostic costs, lengthy processing times, and the need for careful sample storage and maintenance of nucleic acid quality. If an initial RT-PCR test yields a negative result, but subsequent testing confirms the infection, the initial result is deemed a false negative. Statistical reports indicate that approximately 54% of infected patients receive an initial false-negative diagnosis, attributed to factors such as low viral load, early stages of infection, viral evolution, contamination, sample quality, and assay optimization.91 Conversely, false-positive results, which are less common than false negatives, occur when COVID-19-negative patients are incorrectly diagnosed as positive. These errors are often linked to viral load thresholds, protocol-related contamination, sample mishandling, carryover, and data analysis errors.92
RT–LAMP
Loop-mediated isothermal amplification (LAMP) is a single-step nucleic acid amplification technique widely explored for disease diagnosis. It is similar to PCR but does not require a thermocycler, and it is carried out in an isothermal setup. Nucleic acid is incubated with 4–6 target-specific primers (inner, outer, and loop primers) and Bst DNA polymerase at 60–65°C for a single-step amplification and detection, generating ∼109 times amplicons per hour. Real-time amplification can be visualized with the help of DNA binding dyes, turbidity analysis, or pH dye. RT-LAMP merges the idea of reverse transcriptase with LAMP for effective detection of RNA. Reverse transcriptase is added to the RT-LAMP reaction mixture, turning RNA into cDNA and further amplified. RT-LAMP can reportedly detect SARS-CoV-2 RNA within 30 min and is cheaper than RT-PCR.93 AQ-TOP™ COVID-19 Rapid Detection Kit PLUS (Seasun Biomaterials), based on this technique, targets amplification of N and ORF1ab genes in anterior nasal, mid-turbinate nasal, nasopharyngeal and oropharyngeal swabs/aspirates and bronchoalveolar lavage specimens at 60 °C and gives result in 15 min. Clinical evaluation showed 100% positive and negative agreement in 85 individuals, and the kit received emergency use approval on 5th October 2020.94 RT-LAMP had 78% sensitivity in the crude sample whereas 94 % in infected patient purified RNA.97 However, the major challenges include the requirement of experience, assay optimization, and data interpretation. Moreover, under low viral load, RT-LAMP can diagnose the sample as false-negative with a rate of 0.12.95
Metagenomic next-generation sequencing (mNGS)
Upon aligning the RT–PCR diagnosed SARS-CoV-2 cases with the GenBank nucleotide database (2019), CLOMP (Clinically Okay Metagenomic Pipeline) revealed a match between the databases (positive cases and SARS-CoV-associated virus) validating use of mNGS for detection of whole SARS-CoV-2 genome. Unlike PCR, which detects only known viral genes, mNGS can detect the whole genome without any bias and identify alignments with pre-existing viral databases.96 The sensitivity of mNGS was shown by a study where meta-genomic analysis of a SARS-CoV-2 patient showed co-infection with rhinovirus.97 It is highly sensitive and specific, but the high cost of NGS equipment and extensive processing time are the biggest drawbacks of this method.98
CRISPR-based assays
CRISPR (clustered regularly interspaced short palindromic repeats) based approaches use bacterial enzymes (Cas12 and Cas13) which act as a molecular scissor and cut viral RNA at specific locations that are further isothermally amplified and visualized. DETECTR (SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter) couples CRISPR-Cas12 with lateral flow technology to efficiently detect this infection in oropharyngeal and nasopharyngeal swabs. This method is low cost, highly targeted, and sensitive and can give results within an hour.99 Similarly, Sherlock CRISPR SARS- CoV-2 kit (Sherlock BioSciences-US) uses Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) technology which combines the principle of amplification by LAMP and CRISPR to detect ORF1ab and N gene in nasopharyngeal and oro-pharyngeal swabs within 40 min at lowest detection of 675 copies/µl. This technology was approved for emergency use by US FDA on 6th May 2020. The reprogrammable ability of CRISPR allows the diagnosis to stay in line with viral evolution. Conversely, the unavailability of Cas-specific PAM sequence, RNA fragility, and instability makes the approval of these assays challenging.100
Global status of molecular diagnostics development and their regulatory approval
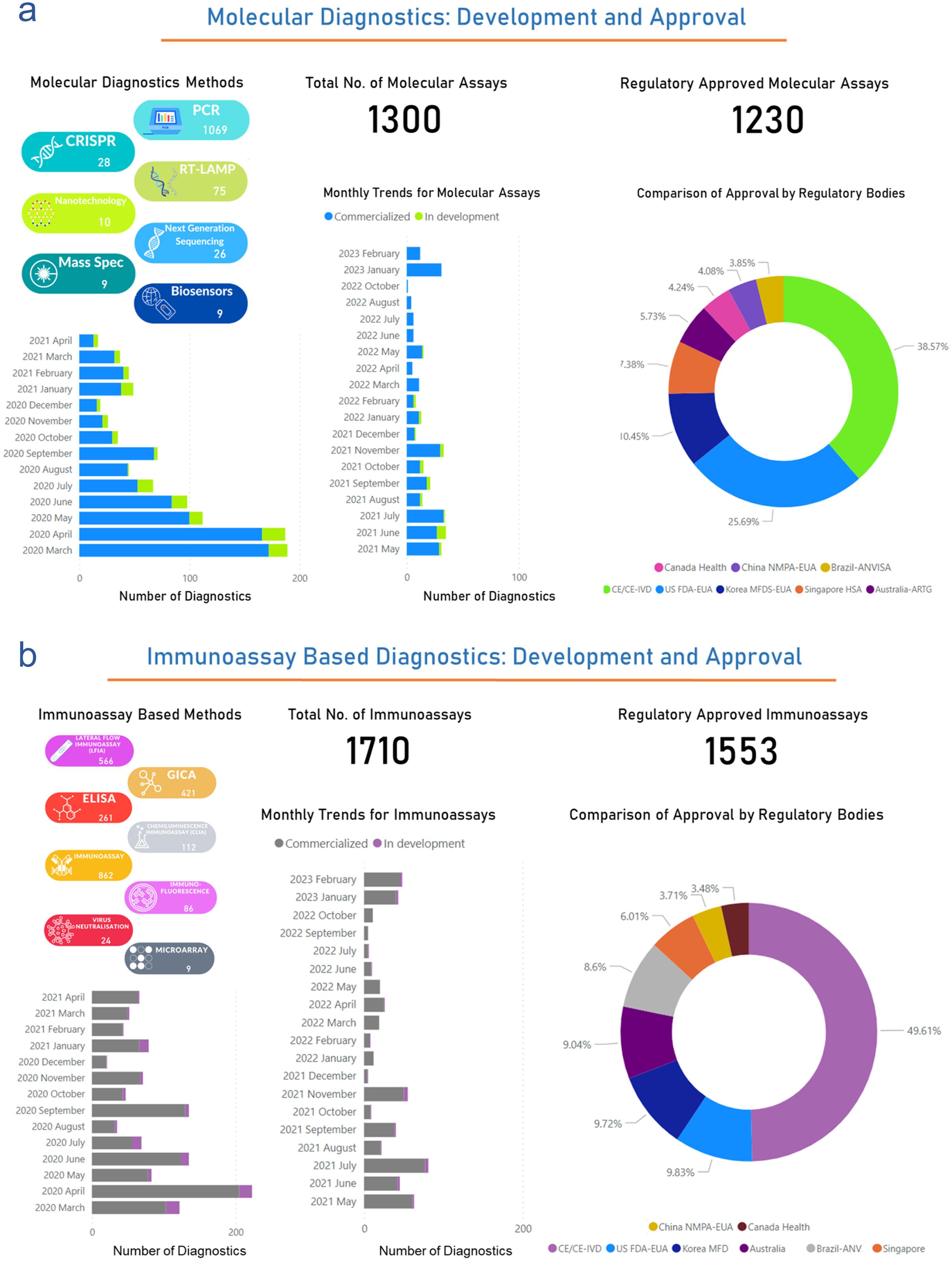
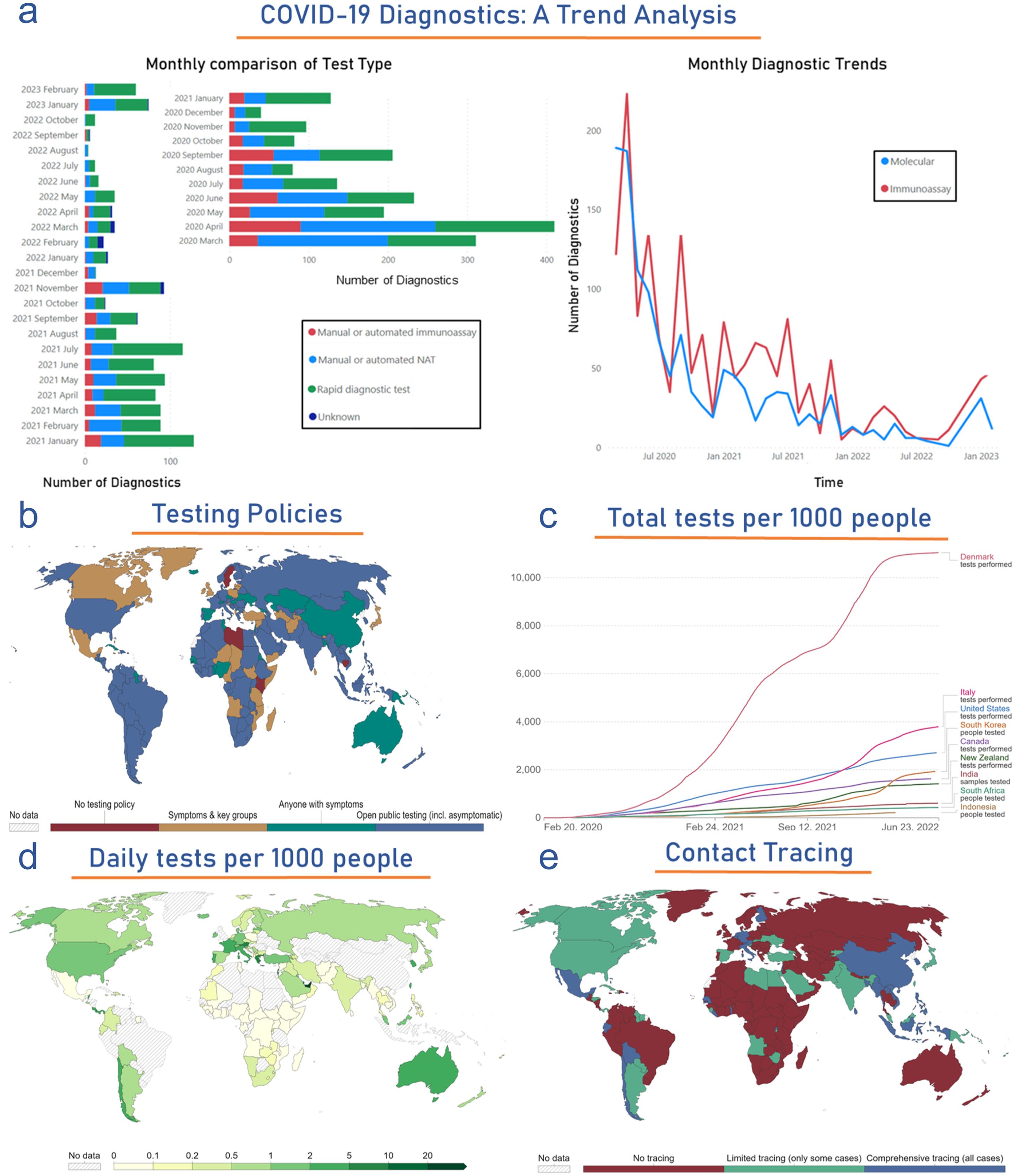
The global status of molecular diagnostics development and their regulatory approval throughout this pandemic has been dynamic and crucial in the fight against the virus. Out of 3,034 diagnostics, 1,300 diagnostics are molecular assay-based tests. However, 1,230/1,300 molecular diagnostics are approved by several regulatory bodies for their clinical use in the diagnosis. Out of 1,230 approved molecular diagnostics, 327 US-FDA-EUA approved (25.69%), 133 Korea MFDS-EUA approved (10.45%), 94 Singapore-HSA approved (7.38%), 73 Australia-ARTG approved (5.73%), 54 Canada Health approved (4.24%), 52 China NMPA-EUA approved (4.08%), 49 Brazil-ANVISA approved (3.85%), and 491 CE/CE-IVD approved (38.57%). Furthermore, since March 2020. The monthly trend of commercialization and the development stage of newer molecular assay-based diagnostics were explained in Figure 4a. Thus, this analysis suggests that a maximum 200 number of molecular diagnostics were reported for their development and commercialization in March and April 2020. This development rate has been reduced but still has a strong side for future pandemics. (http://www.io.nihr.ac.uk/report/covid-19-diagnostics/ )
(a) The left panel of the figure depicts the monthly trends and status of molecular diagnostics in terms of commercial availability or development stage from March 2020 to February 2023. The right panel (pie-chart) shows the percentage of molecular diagnostics approved by the specific regulatory body. (b) The left panel of the figure presents monthly trends and the status of immunoassay-based diagnostics, spanning from March 2020 to February 2023. The right panel (pie chart) highlights the percentage of immunoassay-based diagnostics that have received approval from specific regulatory bodies. Data were obtained from the publicly available dataset http://www.io.nihr.ac.uk/report/covid-19-diagnostics/ . CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; ELISA, Enzyme-Linked Immunosorbent Assay; GICA, Generalized Integrated Circuit Architecture.; PCR, Polymerase Chain Reaction.
In summary, the COVID-19 pandemic has spurred a rapid and collaborative global effort in developing and securing regulatory approval for molecular diagnostics. These tests have been crucial in diagnosing and monitoring the virus, guiding public health responses, and enabling the safe reopening of economies and societies. The focus remains on enhancing the accessibility, accuracy, and speed of testing while adapting to the challenges posed by new variants and evolving testing needs. Continued research and adaptation of diagnostics are essential as the pandemic progresses.
Immunological assays
These assays leverage the antigen-antibody binding affinity to detect either antigenic SARS-CoV-2 proteins or antibodies produced by the host immune system in response to the infection, providing insights into current or past exposure. Compared to NAAT, immunological assays use proteins, which are significantly more stable than RNA, offering portable, straightforward, and cost-effective diagnostic solutions.101
Antigen detection
Several rapid antigen test (RAT) self-diagnosis kits are available. These kits typically include antibodies affixed to a paper strip. When exposed to a sample, the strip binds with any present viral antigen, delivering a visual result within 30–60 min. These strips are sensitive to actively replicating viruses, enabling efficient detection of infections at an early stage. Besides respiratory samples, blood sample testing kits are also available. These kits are user-friendly, fast, and inexpensive, requiring no specialized expertise.90 Currently, 45 kits have been approved by the FDA for emergency use. These kits majorly target N and S proteins of SARS-CoV-2 (https://www.fda.gov ). Some of the antigen detection tests viz., Sofia 2 Flu + SARS Antigen FIA (Quidel Corporation), BD Veritor System for Rapid Detection of SARS-CoV-2 & Flu A+B (BD) and Status COVID-19/Flu A&B (Princeton BioMeditech Corp) are capable of differentiating SARS-CoV-2 and influenza A/B infection by targeting virus-specific N. Brief information regarding the FDA emergency use kits is given in Table 3. In asymptomatic individuals, the sensitivity of NAAT and antigen tests was 80% and 41%, respectively. In symptomatic individuals, the specificity of NAAT and antigen tests was 98% and 99%, respectively. Antigen tests provide higher specificity than NAAT but have low sensitivity. They are highly dependent on viral load and are often found to give false-negative results.98
List of US-FDA-approved commercially available rapid antigen test (RAT) kits
| Kit | Developer | Sample | Tech. | Time (min) | Efficacy |
|---|---|---|---|---|---|
| Antigen: N-protein | |||||
| Sofia 2 Flu + SARS Antigen FIA | Quidel Corporation | NS, NPS | LFI | 15 | Sensitivity-95.2%; Specificity-100% |
| Clip COVID Rapid Antigen Test | Luminostics, Inc. | ANS | LFI | 30 | LOD: 0.88 102 TCID50 /mL |
| Ellume COVID-19 Home Test* | Ellume Limited | MTNS | LFI | 15 | Accuracy: 96% |
| QuickVue At-Home COVID-19 Test* | Quidel Corporation | ANS | LFI | 10 | LOD: 1.91 × 104 TCID50 /mL |
| BD Veritor System for Rapid Detection of SARS-CoV-2 & Flu A+B | Becton, Dickinson and Company (BD) | ANS | CDI | 15 | LOD: 2.8 × 102 TCID50 /mL |
| Omnia SARS-CoV-2 Antigen Test | Qorvo Biotechnologies, LLC. | ANS | BAWB | 20 | LOD: 200 TCID50 /mL |
| Sofia SARS Antigen FIA | Quidel Corporation | ANS | LFI | 15 | Sensitivity-96.7%; Specificity- 100% |
| ellume.lab COVID Antigen Test | Ellume Limited | MTNS | LFI | 15 | LOD: 7.16 × 103 TCID50 /mL |
| LIAISON SARS-CoV-2 Ag | DiaSorin, Inc | NPS | CLI | 120 | LOD: 300 TCID50 /mL |
| QIAreach SARS-CoV-2 Antigen Test | QIAGEN GmbH | NPS, ANS | LFI | 2–15 | ANS: sensitivity-85%, specificity-99.05%; NPS: sensitivity-80.65%, specificity-98.31% |
| SCoV-2 Ag Detect Rapid Test | InBios International, Inc. | ANS | LFI | 20 | Sensitivity-86.67%; Specificity-100% |
| NIDS COVID-19 Antigen Rapid Test Kit | ANP Technologies, Inc. | MTNS | LFI | 15 | LOD: 311 TCID50 /mL |
| SPERA COVID-19 Ag Test | Xtrava Health | ANS | LFI | 15–30 | LOD: 1.56 × 103 TCID50 /mL |
| Flowflex COVID-19 Antigen Home Test* | ACON Laboratories, Inc | ANS | LFI | 15 | Sensitivity- 93%; specificity- 100% |
| QuickVue At-Home OTC COVID-19 Test* | Quidel Corporation | ANS | LFI | 10 | LOD: 1.91 × 104 TCID50 /mL |
| Status COVID-19/Flu A&B | Princeton BioMeditech Corp. | ANS, NPS | LFI | 15 | LOD: 2.7 × 103 TCID50 /mL |
| LumiraDx SARS-CoV-2 Ag Test | LumiraDx UK Ltd. | ANS, NPS | MI | 12 | Sensitivity-97.6%; specificity-96.6% |
| QuickVue SARS Antigen Test | Quidel Corporation | ANS | LFI | 10 | LOD: 1.51 × 104 TCID50 /mL |
| VITROS Immunodiagnostic Products SARS-CoV-2 Antigen Reagent Pack | Ortho Clinical Diagnostics, Inc. | ANS, NPS | CLI | 48 | LOD: 5 × 102-2.7 × 103 TCID50 /mL |
| GenBody COVID-19 Ag | GenBody Inc. | ANS, NPS | LFI | 15–20 | LOD: 1.11 × 102 TCID50 /mL |
| BD Veritor At-Home COVID-19 Test* | BD | ANS | LFI | 15 | LOD: 1.87 × 102 TCID50 /mL |
| CareStart COVID-19 Antigen | Access Bio, Inc | ANS, NPS | LFI | 10 | LOD: 8 × 102 TCID50 /mL |
| BD Veritor System for Rapid Detection of SARS-CoV-2 | BD | ANS | CDI | 15 | LOD: 1.4 × 102 TCID50 /mL |
| Sienna-Clarity COVID-19 Antigen Rapid Test Cassette | Salofa Oy | NPS | LFI | 10 | LOD: 1.25 × 103 TCID50 /mL |
| Simoa SARS-CoV-2 N Protein Antigen Test | Quanterix Corporation | ANS, NPS | PMI | 80 | LOD: 0.29 TCID50 /mL |
| iHealth COVID-19 Antigen Rapid Test* | iHealth Labs, Inc | ANS | LFI | 15 | LOD: 20 × 103 TCID50 /mL |
| COVID-19 At-Home Test* | SD Biosensor, Inc | ANS | LFI | 15–30 | Sensitivity-94.94%; Specificity-100% |
| BinaxNOW COVID-19 Antigen Self-Test* | Abbott Diagnostics Scarborough, Inc. | ANS | LFI | 15 | LOD: 140.6 TCID50 /mL |
| BinaxNOW COVID-19 Ag Card 2 Home Test* | Abbott Diagnostics Scarborough, Inc. | ANS | LFI | 15 | LOD: 140.6 TCID50 /mL |
| INDICAID COVID-19 Rapid Antigen Test | PHASE Scientific International, Ltd. | ANS | LFI | 20 | LOD: 140 TCID50 /mL |
| iHealth COVID-19 Antigen Rapid Test Pro | iHealth Labs, Inc | ANS | LFI | 15 | LOD: 20 × 103 TCID50 /mL |
| MaximBio ClearDetect COVID-19 Antigen Home Test* | Maxim Biomedical, Inc. | ANS | LFI | 15 | Sensitivity-86.9%; Specificity-98.9% |
| CareStart COVID-19 Antigen Home Test* | Access Bio, Inc. | ANS | LFI | 10 | LOD: 800 TCID50 /mL |
| SCoV-2 Ag Detect Rapid Self-Test* | InBios International Inc | ANS | LFI | 25 | LOD: 6.3 × 103 TCID50 /mL |
| InteliSwab COVID-19 Rapid Test Rx | OraSure Technologies, Inc. | ANS | LFI | 30–40 | LOD: 2.5 × 102 TCID50 /mL |
| InteliSwab COVID-19 Rapid Test* | OraSure Technologies, Inc | ANS | LFI | 30–40 | LOD: 2.5 × 102 TCID50 /mL |
| InteliSwab COVID-19 Rapid Test Pro | OraSure Technologies, Inc | ANS | LFI | 30–40 | LOD: 2.5 × 102 TCID50 /mL |
| Nano-Check COVID-19 Antigen Test | Nano-Ditech Corp | NPS | LFI | 10–15 | Sensitivity-90.32%; Specificity-100.0% |
| BinaxNOW COVID-19 Ag Card | Abbott Diagnostics Scarborough, Inc. | ANS | LFI | 15 | LOD: 140.6 TCID50 /mL |
| BinaxNOW COVID-19 Ag Card Home Test* | Abbott Diagnostics Scarborough, Inc | ANS | LFI | 15 | LOD: 140.6 TCID50 /mL |
| BinaxNOW COVID-19 Ag 2 Card | Abbott Diagnostics Scarborough, Inc | ANS | LFI | 15 | LOD: 140.6 TCID50 /mL |
| CLINITEST Rapid COVID-19 Antigen Self-Test | Siemens Healthineers | ANS, NPS | LFI | 15 | ANS: sensitivity-97.25%, specificity-100%; NPS: sensitivity-98.32%, specificity-99.6 % |
| Antigen: S (RBD) Protein | |||||
| Sampinute COVID-19 Antigen MIA | Celltrion USA, Inc | NPS | MESI | 10 | Sensitivity-94.4%; Specificity-100% |
| Antigen: N + S (RBD) protein | |||||
| Celltrion DiaTrust COVID-19 Ag Home Test* | Celltrion USA, Inc | MTNS | LFI | 15 | Sensitivity-86.7%; Specificity-99.8% |
| Celltrion DiaTrust COVID-19 Ag Rapid Test | Celltrion USA, Inc | MTNS | LFI | 15 | LOD: 3.2 × 101 TCID50 /mL |
Serological analysis/Antibody detection
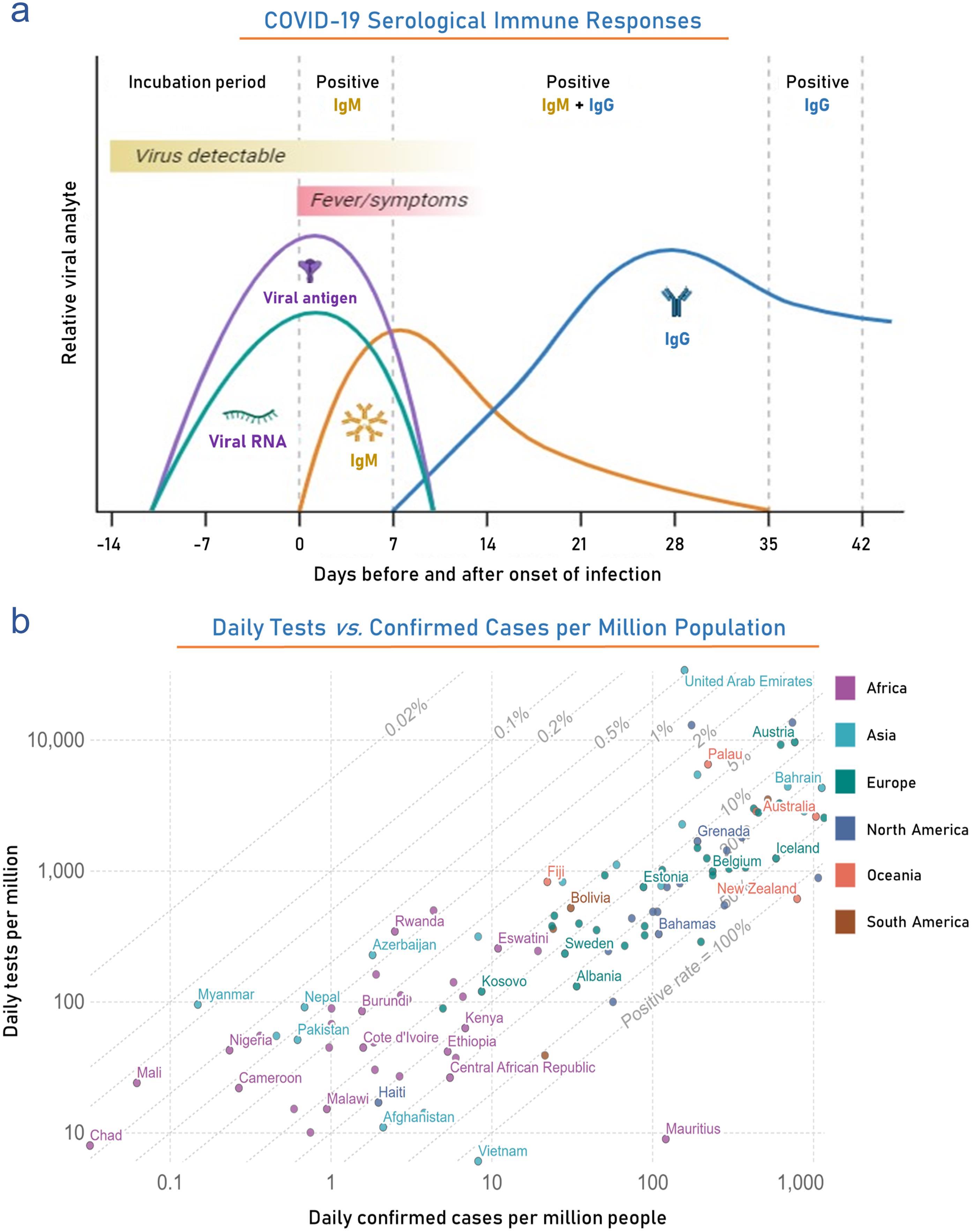
Serological tests can diagnose current or past infection by detecting antibodies in patient sera (Fig. 3b). Specific antibody development takes around a week, so sensitivity towards early or acute infection is very low. Infection history and initial exposure date can be estimated by analyzing the seroconversion of different immunoglobins. IgM becomes detectable after 1 week of infection, peaking at weak 2 and then coming down to basal level, whereas IgG, detected after 1 week, remains high for a prolonged period. Peptide-based luminescent immunoassay, ELISA, immunochromatographic assay, and lateral flow immunoassay are some of the well-explored antibody detection techniques.99 A list of serological and antibody-based induced adaptive immune response tests approved by US-FDA are given in Table 4.102 Among these Elecsys Anti-SARS-CoV-2 S, an electro-chemiluminescence immunoassay developed by Roche Diagnostics and approved for emergency use by the FDA in November 2020, can identify the presence of active immune response, an indication of past or current SARS-CoV-2 S infection. It can detect and partially quantify anti-RBD antibodies (an immunological response of SARS-CoV-2 S) in human serum and plasma by incubating the sample with dual antigens, SARS-CoV-2 S-RBD recombinant antigen tagged with biotin and ruthenium. Analysis of 5,272 samples showed 99.81 % Specificity, and 204 samples analyzed after PCR detection had sensitivity in the range of 65.5 % (0–6 days) to 100 % (≥14 days). A chemiluminescent immunoassay, Atellica IM SARS-CoV-2 IgG (sCOVG), developed by Siemens Healthcare Diagnostics Inc., can detect IgG formed against SARS-CoV-2 in human serum/plasma. This kit contains an Atellica IM sCOVG DIL solution and biotinylated SARS-CoV-2 recombinant antigens coated Solid Phase Reagent run on Atellica IM Analyzer. Clinical data reports the sensitivity ranged from 50% (0–7 days) to 95.58% (≥ 15) post PCR detection in 711 participants whereas in 1993 participant sensitivity 99.9%.
US-FDA and EUA-approved commercially available serological test kits*
| Kit name | Kit #Cat. No. | Test/kit | Developer | Detection |
|---|---|---|---|---|
| RapCov™ | A-RAPCOV01 | 25 | Advaite | IgG |
| CovAb | 2039 | 50 | Diabetomics | IgG/IgA/IgM |
| ADEXUSDx COVID-19 Test | 8075 | 50 | NOW Diagnostics | Total Ig |
| SGTi-flex COVID-19 | COGT025E, COGT005E | 25, 5 | Sugentech | IgG |
| TBG SARS-CoV-2 | 20010 | 25 | TBG Biotech | IgG/IgM |
| ACON | L031-11711 | 25 | ACON Laboratories | |
| Sienna-Clarity COVIBLOCK COVID-19 | CD-COV19CW/102223/102224 | 20 | Salofa Oy | |
| Telepoint | – | 25 | Xiamen Biotime Biotech | |
| BIOTIME | – | 25 | ||
| RightSign™ | – | 20 | Hangzhou Biotest Biotech | |
| CoronaCHEK | – | 25 | Hangzhou Biotest Biotech | |
| Premier Biotech COVID-19 Rapid Test | – | – | ||
| LYHER | 303002 | 40 | Hangzhou Laihe Biotech. | |
| QUICKKIT | – | – | Hangzhou Laihe Biotech | |
| COVID-19 rapid Test | GCCOV-402a | 25 | Healgen Scientific Limited Liability | |
| 2019-nCov Ab Test | YF319C | 20 | Innovita Biological Tech. | |
| Orawell Rapid Test | – | – | Jiangsu Well Biotech | |
| INDICAID COVID-19 | – | 25 | Jiangsu Well Biotech | |
| Rapid COVID-19 | – | 25 | Megna Health | |
| MidaSpotTM COVID-19 | NBPC-0007 | 25 | Nirmidas Biotech | |
| Nirmidas COVID-19 | NBPC-0001-xx | 20 | ||
| Assure | COV-W23M | – | Assure Tech. | |
| Ecotest | – | 2, 5 | ||
| Fastep | – | – | ||
| Wantai SARS-CoV-2 Ab Rapid Test kit | WJ-2710, WJ-2750 | 10 50 | Beijing Wantai Biological Pharmacy Enterprise | |
| Tell Me Fast | B251C | 25 | Biocan Diagnostics | |
| SARS-CoV-2 Ab Test | RTA0203 | 25 | Biohit Healthcare |
Global status of immunoassays based-diagnostics development and their regulatory approval
Methods like lateral flow immunoassays, chemiluminescence based immunoassays, GICA, ELISA, immunofluorescence based, and microarray-based several serological kits and assays are developed for their clinical useThe global status of diagnostic development and its regulatory approval has been a pivotal element in the worldwide response to the pandemic. Our analysis indicates that of the 3,034 diagnostics evaluated, approximately 1,710 are immunoassay-based tests. Of these, 1,553 have garnered clinical approval from various regulatory entities for diagnostic purposes. The distribution of approvals among these immunoassay-based diagnostics is as follows: 153 (approximately 9.8%) by the US FDA under EUA, 152 (approximately 9.72%) by the Korea MFDS under EUA, 94 (approximately 6.01%) by Singapore’s HAS, 141 (approximately 9.04%) by Australia’s ARTG, 53 (approximately 3.48%) by Health Canada, 56 (approximately 3.71%) by China’s NMPA under EUA, 134 (approximately 8.6%) by Brazil’s ANVISA, and 770 (approximately 49.61%) have received CE or CE-IVD approval. Starting in March 2020, Figure 4b illustrates a monthly trend in the commercialization and developmental stages of new immunoassay-based diagnostics. This analysis reveals that the peak, with approximately 220 diagnostics, occurred in April 2020 during the height of the pandemic. Although the rate of development has since declined, it remains significant, suggesting a sustained trajectory that will be crucial for future pandemic preparedness. (http://www.io.nihr.ac.uk/report/covid-19-diagnostics/ )
In summary, the development and regulatory approval of these diagnostics have been vital in the global response to this pandemic. These tests have played a crucial role in diagnosing past infections, conducting seroprevalence studies, and monitoring vaccine responses. However, the landscape has seen variations in test performance and adaptation efforts to address the evolving nature of the virus. International collaboration and stringent regulatory oversight have been essential components of this effort.
Imaging examination
Besides investigating SARS-CoV-2 biological components, imaging techniques, such as CT-scan, X-Ray, MRI, and lung ultrasonography, can diagnose COVID-19 based on the anatomical changes in the respiratory tract and lungs. These examinations can effectively identify lung collapse, pleural effusions, pneumothorax, and pulmonary edema associated with severe COVID-19 infection. Chest X-ray (CXR) shows 69% sensitivity against COVID-19 by detecting hazy opacities, peripherally, and bilateral lower zone consolidation. CT scans can show septal thickening and ground-glass consolidated opacities.103 However, the abnormalities are not limited to SARS-CoV-2 specific infection but can also result from underlying disease.98 CT images of most COVID-19 patients show similar patterns, such as bilateral patchy distribution, ground glass-like opacity, and sometimes circular-shaped peripheral distribution in the lungs.104 The bilateral and frosted glass-like opacity observed in chest CT scans is a characteristic finding in COVID-19, indicating diffuse alveolar damage and inflammatory changes within the lungs, leading to impaired gas exchange.105–107 Additionally, a new Cas13-based SHERLOCK technology can also be utilized to detect SARS-CoV-2 infection. In this system, the Cas13 enzyme targets and cleaves the RNAs, which were used for amplifying a reporter signal in diagnostic tests.108 Taken together, other technologies such as immune chromatography, colloidal gold, and other associative biotechnologies are in progress.
Comparative analysis of developed methods
Each existing approach for identifying SARS-CoV-2 has its designated applications, but they are all burdened by their inherent shortcomings. As a result, ongoing research endeavors continue to search for alternative detection methods that can enhance sensitivity, precision, and detection speed. Several diagnostic techniques have virus detection capability at specific stages. In the following section, we offer a succinct assessment of the previously mentioned techniques and introduce a range of potentially auspicious diagnostic methods for COVID-19, focusing on addressing the current deficiencies in detection capabilities (Fig. 5a).109
(a) Side-bar plot compares various diagnostic methods in the monthly COVID-19 cases diagnosed from March 2020 to February 2023. Meanwhile, the right-side scatter plot compares the molecular and immunoassay-based diagnosed COVID-19 cases throughout the pandemic. The figure shows global statistics of (b) testing policies, (c) the Maximum number of tests performed per thousand people in top selected countries, (d) Daily global testing, and (e) Level of contact tracing. Data were obtained from the analysis of publicly available datasets http://www.io.nihr.ac.uk/report/covid-19-diagnostics/ and https://ourworldindata.org/coronavirus . VOC, Volatile Organic Compounds.
The primary drawback of PCR-based methods is their constrained sensitivity, leading to potential false negatives in early infection. This method depends upon supplementary clinical observation and medical history. Furthermore, this method requires specialized facilities, equipment, and trained personnel, posing challenges in smaller or rural healthcare facilities. Additionally, due to limited reagent availability, PCR-based tests often face shortages. Moreover, these tests are invasive and time-consuming, with hours-long result times. They can detect the virus even in the early stages of infection when the viral load is low. Furthermore, PCR-based methods are designed to detect the presence of SARS-CoV-2 but cannot track asymptomatic infections and recoveries.109,110 Serology tests can identify individuals exposed to COVID-19, offering a significant advantage by detecting recent and ongoing infections. This capability makes serology tests a valuable tool for assessing the true prevalence of the virus within a specific population. Additionally, they can provide insights into the infection stage by measuring the antibody level in the specimen. However, it is crucial to recognize that serology tests do not directly detect the virus; instead, they identify antibodies produced in response to the virus. As such, they share a common limitation with PCR-based methods, potentially yielding false-negative results, especially in the early stages of infection.109
In contrast, chest CT scans demonstrate superior sensitivity compared to both serology and PCR-based methods, particularly in the early stages of infection. However, implementing chest CT scans requires expensive equipment and skilled operators. Furthermore, the radiographic abnormalities observed in COVID-19 cases can resemble those of other viral pneumonias, meaning that chest CT scans cannot definitively confirm COVID-19 infection.109 Chest X-ray machines are economical and widely accessible substitutes for CT scans, but they have limitations in sensitivity and specificity compared to CT scans. Advances in AI, including machine learning and deep learning, enhance their diagnostic capabilities. Computer-aided diagnosis systems enable the use of chest X-rays for COVID-19 diagnosis. This makes chest X-rays a promising tool, especially in resource-limited regions, such as low to medium-income countries.109,111
Variant specific detection
The continuous evolution of SARS-CoV-2 demands up-to-date diagnostic modalities. Identification of VOCs (Alpha, Beta, Gamma, Delta, and Omicron variants) is an essential prerequisite of therapy development. The most reliable way of variant detection is the whole viral genome or at least S-gene sequencing. Nevertheless, instrumental unavailability, complexity, and high expertise requirements make sequencing difficult for early infection diagnosis, variant contact tracing, and prevalence calculation. Multiplex RT-PCR of the Alpha variant gives signals for nucleocapsid and ORF1 genes but not for S-gene, indicating S-gene target failure. This RT-PCR result pattern can be used for Alpha variant diagnosis as it is not present in Beta and delta variants. However, this target failure is not limited to the Alpha variant; it could also be found in other mutated forms like Omicron. A fast variant diagnosis assay, SNP targeted RT-PCR, can detect Alpha variant specific mutation like spike HV69-70del and N501Y in less than an hour.112 University Hospital Geneva identified Omicron by partial Sanger sequencing of two S gene regions followed by RT-PCR. Thermo Fisher TaqPath identified ΔH69/V70 of Omicron by S-gene target failure.113 TIB MolBiol did an RT-PCR melting curve analysis to identify S371L/S373P, ins214EPE, and E484A of the same variant.114 The alpha variant was successfully detected in wastewater by allele-specific RT-qPCR targeting Y144del, HV69/70del, and A570D mutations of the specific variant.115 A similar study showed that primer based on 21,724–21,828 of alpha variant and 22,243–22,331 bp of beta variant, S gene led to efficient detection of the variants in wastewater by RT-qPCR.116 Rapid antigen tests (RAT) can detect most variants, but their differentiation is not yet possible due to the low sensitivity of RAT. As most of the antigen-based assays target nucleocapsid, the major mutation in the spike gene of VOCs does not significantly affect RAT sensitivity and efficacy, making this approach favorable for early diagnosis and contact tracing.117 The detection potential of the Sure Status COVID-19 Antigen Card Test (Premier Medical Corporation) and Flowflex SARS-CoV-2 Antigen Rapid Test (ACON Laboratories) against different VOCs showed that Sure Status COVID-19 Antigen Card Test could efficiently diagnose alpha, beta, and gamma variants, whereas Flowflex SARS-CoV-2 Antigen Rapid Test had major sensitivity for delta variant.118 A RAT kit by E25Bio, Inc., Cambridge, MA, and Perkin Elmer, Waltham, MA, targeting the N protein showed high sensitivity of alpha and beta variants followed by omicron and delta. The low sensitivity of delta could result from a mutation in the N gene.119 Abbott antigen, serological, and molecular test kits could also detect alpha, beta, gamma, and delta variants.120
Serological study is essential for determining the risk associated with the emergence of different variants on transmissibility, mortality, and morbidity in vaccinated and pre-infected candidates and vaccine escape potential. To estimate the defensive ability of humoral antibodies induced by infection and vaccine against the new variants, proper analysis of virus neutralization capacity in plasma and/or sera of candidates is essential. Pseudovirus neutralization assay, microneutralization, and plaque reduction neutralization (PRNT) are some assays developed to find neutralization capacity.121–123 As an international standard, WHO recommends using high titer reference serum and WHO International Antibody Standard (WHO IS)/NIBSC working reagent for neutralization assays.112 The neutralization capacity of the Beta variant was analyzed by live-virus neutralization assay in the plasma of infected individuals from two waves of COVID-19 in South Africa, where the second wave was predominated by the Beta variant. The beta variant was neutralized efficiently with the plasma of the second wave infected patient, but upon neutralization with the first phase plasma, the efficacy was reduced by 15.1 folds. However, when the first-wave non-VOC variant was neutralized with second-phase plasma, only 2.3-fold decreases were observed. This indicates that a vaccine based on VOC may elicitate immunity against other variants.124 Delta variant (B.1.617.1, B.1.617.2, and B.1.351) neutralization was studied in individuals vaccinated with ChAdOx1 (Oxford/AstraZeneca) and BNT162b2 (Pfizer/BioNTech). B.1.617.1, B.1.617.2, and B.1.351 reduced neutralization by 4.31, 5.11, and 6.29 folds in vaccinated candidates, and after dual dose vaccination by BNT162b2, the reduction was increased to 7.77, 11.30 and 9.56 folds. This shows that two doses of vaccines are essential for defense against different variants.125 Omicron (B.1.1.529) pseudovirus neutralization assay reduced neutralizing antibody titer by 45 folds. Infected and vaccinated individuals showed prominent cross-neutralization with a 5-fold potency reduction.126
Global status of VOC’s diagnostics development, sequences identification, and regulatory approval
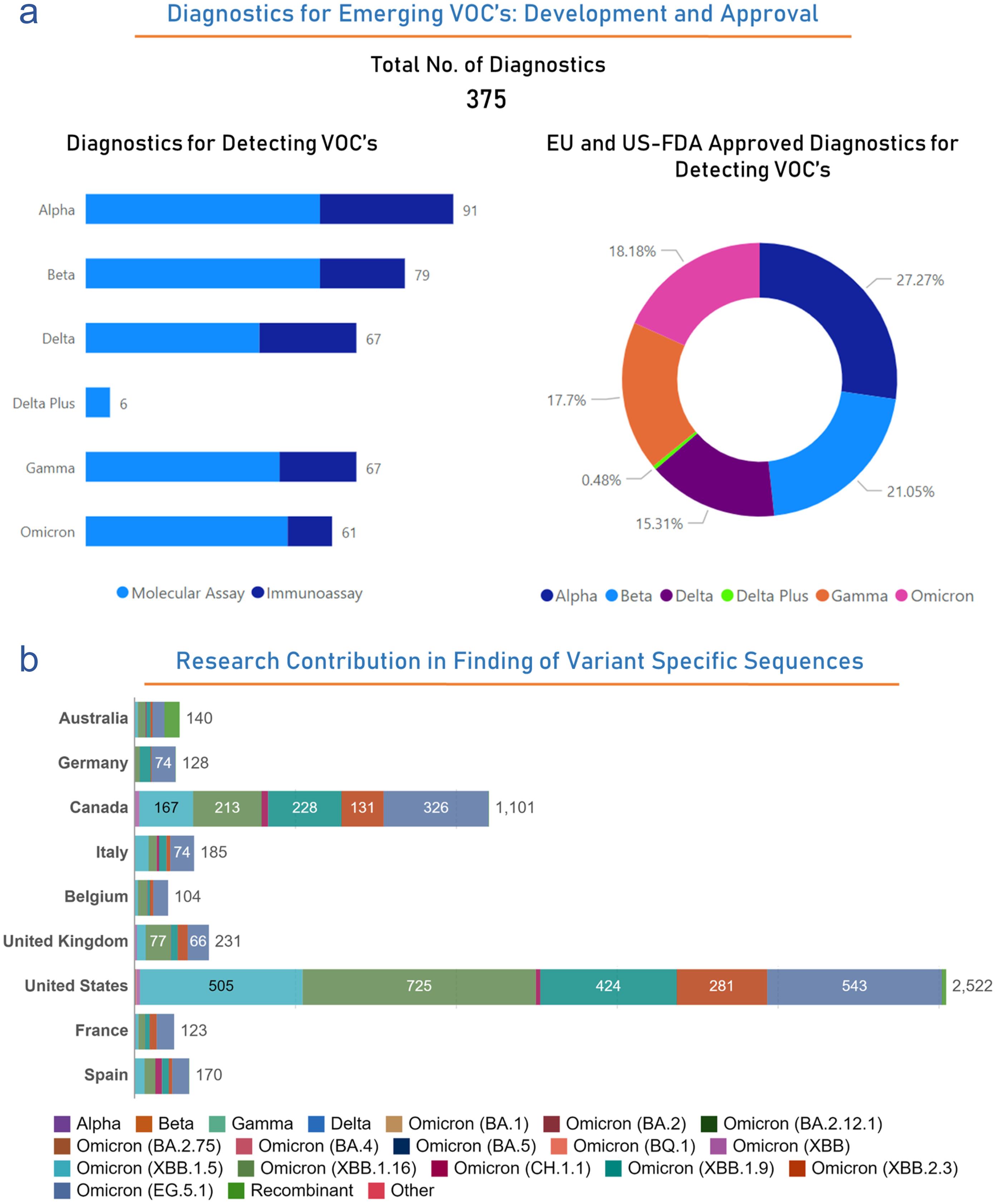
Throughout this pandemic, the detection and monitoring of VOCs have been critical in understanding the evolution of the virus and adapting public health responses. Based on our analysis, out of the 3,034 diagnostics, 375 have received clinical approval from various regulatory bodies to detect several VOCs (http://www.io.nihr.ac.uk/report/covid-19-diagnostics/ ). Among the 375 approved diagnostics, the breakdown approvals by regulatory bodies for specific variant detection is as follows: 91 diagnostics for Alpha (Molecular: 58; Immunnoassay: 33), 79 diagnostics for Beta (Molecular: 58; Immunnoassay: 21), 67 diagnostics for Delta (Molecular: 43; Immunnoassay: 24), 6 diagnostics for Delta Plus (Molecular: 6; Immunnoassay: 0), 67 diagnostics for Gamma (Molecular: 48; Immunnoassay: 19), and 61 diagnostics against Omicron variant (Molecular: 50; Immunnoassay: 11). Above mentioned diagnostics are also approved by several regulatory bodies for their clinical use mentioned in Figure 6a. Several countries have played roles in determining newly emerged variants by determining their sequences. Countries have played a significant role in determining the maximum number of sequences of several variants throughout the pandemic (https://ourworldindata.org/coronavirus ). The United States has identified 2,522, Canada has identified 1,101, the United Kingdom has identified 231, Australia has identified 140, Germany has identified 128, Italy has identified 185, Belgium has identified 104, France has identified 1,101, and Spain has identified 170 sequences of several variants of SARS-CoV-2 throughout this pandemic (Fig. 6b). In summary, the global response to VOCs during the pandemic has involved the development of specialized diagnostics, regulatory approvals, international collaboration, and adjustments to public health measures. Monitoring and adapting to the evolving nature of the virus, particularly through genomic sequencing, have been essential in managing the pandemic and protecting public health.
(a) The left side-bar plot illustrates the number of diagnostics effective against several VOCs, while the right side pie diagram shows the EU and US-FDA-approved number of diagnostics against VOCs. (b) Number of VOC sequences identified by top-most countries. United States is leading with the identification of 2522 VOC sequences, to date. Data were obtained from the publicly available datasets http://www.io.nihr.ac.uk/report/covid-19-diagnostics/ and https://ourworldindata.org/coronavirus . VOC, Volatile Organic Compounds.
Global COVID-19 diagnostics: Shortage and production challenges
The global development of both molecular and immunoassay-based diagnostics has seen significant fluctuations throughout the pandemic. Our analysis reveals that in April 2020 alone, over 400 diagnostics were developed, showing the urgent demand for these tools. Initially, this surge helped alleviate the diagnostic shortages, providing critical support to healthcare systems globally. However, the emergence of new virus variants has highlighted the ongoing need for a steady supply of diagnostics that can adapt to evolving mutations and updated protocols. Both companies and research institutions have been crucial in advancing the development and availability of these vital resources.
The pandemic has exerted unprecedented demands on diagnostic testing worldwide. As the virus spread rapidly, precise and accessible diagnostics became crucial in managing the pandemic. However, this increased demand led to significant challenges in fulfilling testing needs, causing a worldwide shortfall in diagnostics. The rapid proliferation of the virus necessitated mass testing to identify and isolate infected individuals, particularly those asymptomatic or pre-symptomatic. Disruptions in the global supply chain affected the availability of critical testing materials, such as reagents, swabs, and kits, due to heightened demand and interruptions in manufacturing and transport. This resulted in shortages of essential components.
Molecular diagnostics like PCR tests involving intricate manufacturing processes and specialized equipment required scaling up production—a process that demanded time and resources. Additionally, regulatory approvals and quality control measures further delayed the production and distribution of these diagnostics. The requirement for Emergency Use Authorizations (EUAs) or other regulatory clearances, coupled with the varying sensitivity and specificity of the tests, introduced uncertainties about their appropriateness for different scenarios, complicating testing strategies. The appearance of new variants necessitated continuous reevaluation and adjustment of diagnostic tests to maintain their effectiveness. Ensuring equitable access to diagnostics, particularly in low- and middle-income countries, has been a persistent global challenge, with disparities in access exacerbating the diagnostic shortage.127,128
Researchers and diagnostic companies worldwide worked tirelessly to develop and improve testing technologies, including faster and more accessible options. Governments and organizations worked to stabilize the supply chain by increasing production capacity, diversifying suppliers, and addressing logistical challenges. Regulatory agencies introduced expedited approval processes, such as EUAs, to accelerate the availability of diagnostic tests. Global collaboration and information sharing facilitated the development and distribution of tests and helped address disparities in access. Manufacturers scaled up production of diagnostic components and tests to meet growing demand. Ongoing innovations, such as developing point-of-care (POC) tests and self-administered home tests, aimed to make testing more accessible and convenient.
In conclusion, the global shortage of diagnostics was a multifaceted challenge driven by high demand, supply chain disruptions, and regulatory complexities. However, the global response included efforts to expand production, streamline approvals, and promote international collaboration to ensure equitable access to testing.129 These efforts have been crucial in managing the pandemic and preparing for future challenges.
Distinctive features of SARS-CoV-2: An impact on diagnostic approaches
SARS-CoV-2 exhibits unique trait settings apart from seasonal coronaviruses and SARS-CoV, helping significantly shape the testing approaches.
Viral transmission by asymptomatic and pre-symptomatic individuals
High viral loads in the nasal passages are detectable among infected individuals, regardless of their clinical presentation, which leads to classifying this infection as asymptomatic, pre-symptomatic, or symptomatic.130 This characteristic shows the inadequacy of relying solely on symptom-based testing to curb the virus’s spread, emphasizing the urgency of community-based testing. Particularly worrisome are healthcare workers and individuals in residential care homes for those aged 65 and older, as they face a heightened risk of unintentionally transmitting SARS-CoV-2 to both their own families and those under their care (Fig. 5b).131
Period of infection
Data from 113 studies across 17 countries reveal that SARS-CoV-2 RNA can be detected six days before symptoms, peaks around symptom onset, and typically disappears from upper respiratory samples within two weeks. Lower respiratory samples may have higher, delayed, and more persistent viral loads.132 Research utilizing viral cultures indicates that while patients may test RNA-positive for a week post-symptoms, viable virus isolation becomes unlikely after 9 days post-symptom onset. This suggests infectivity mainly spans 2–3 days before to 8 days after symptoms. The presence of RNA-positive culture-negative samples suggests the potential presence of genomic fragments rather than ongoing viral replication.133–135
VOCs
SARS-CoV-2, An RNA virus, due to its inherent instability, frequently undergoes mutations during replication in human cells, leading to the formation of variants. Some of these variants may acquire advantageous traits, such as increased transmissibility. The WHO classifies VOCs based on their significant impact on global public health, including heightened transmissibility or resistance to public health interventions, such as diagnostics, vaccines, and therapeutics. The CDC and the European CDC have developed comparable operational criteria to monitor the emergence of these problematic mutations, which could potentially pose global or regional health threats. In vitro studies have demonstrated that certain VOCs can evade neutralizing antibodies produced through natural infection or vaccination.136 The US FDA supervises molecular assay performance under its EUA list, specifically assessing reduced sensitivity and false negatives linked to VOCs. Antigen tests directed at the SARS-CoV-2 N protein exhibit lower vulnerability to VOC-related issues. A monitoring dashboard by the Program for Technology in Health tracks test validation against VOCs conducted by various companies.79,137–139
Host immune responses
Over a year into the pandemic, our comprehension of the immune reaction to this infection remains unfinished. Data indicate that humoral and cellular immune responses begin 1–2 weeks post-symptoms, with antibodies targeting viral surface proteins and cellular responses encompassing a broader range of viral proteins.140 Following this infection, IgM and IgG antibody development occurs earlier than in other viral infections, peaking at day 11–14 post-symptom onset. Unlike other infections, IgM and IgG antibodies typically emerge simultaneously, enabling the use of IgM antibody tests alongside molecular tests for improved case detection in late-presenting individuals and contact tracing.141 Viral dynamics and antibody response during symptomatic SARS-CoV-2 infection are shown in (Fig. 7a).
(a) Figure illustrates the presence and detection of antibodies in the host. IgM provides initial defense in viral infections, followed by adaptive IgG responses for immunity and memory. Testing COVID-19 IgM and IgG is effective for rapid diagnosis. IgM suggests recent exposure, and IgG indicates a later infection stage, offering infection insights. (b) Graphical representation of continent-wise positive test rate of COVID-19 diagnosed patients. Data were obtained from the publicly available dataset https://ourworldindata.org/coronavirus , and some figure elements were created using Biorender (https://www.biorender.com/ ).
Immunity persistence and the risk of reinfection
Respiratory virus reinfection is frequent, primarily due to waning immunity. The reinfection is characterized by recurrent symptoms and a positive PCR test more than 90 days post-initial infection, validated by exposure history or sequencing.142 In Denmark, a study found 80.5% protection over 7 months, declining to 47.1% for individuals aged 65 and above. Evidence indicates that SARS-CoV-2 antibodies might not grant enduring immunity, showing the need for vigilant reinfection monitoring amid emerging variants.143
Neutralizing antibodies, vaccination, variants, and immune protection
IgG antibodies against SARS-CoV-2 S and N proteins correlate with in vitro neutralization. Elevated IgG in severe cases does not guarantee protection. Neutralizing antibody assays measure in vitro pathogen inactivation, needing secure labs for culturing.144 SARS-CoV-2 neutralization assays using pseudo-viruses offer improved safety. It is crucial to prioritize developing and validating neutralization assays for monitoring variant strains using sera from natural infections or vaccinations.145 A clear protective antibody threshold has not been established, likely influenced by viral variants, viral loads, and other factors. Cellular immune responses play a significant role; in vivo protection correlates are unclear. Therefore, antibody tests should not guide personal or occupational exposure decisions or personal protection. Vaccine-induced immunity primarily targets the S protein. Analyses of sera from vaccinated or naturally infected individuals show limited neutralization against beta (B.1.351) and gamma (P.1) variants, mainly due to spike receptor-binding domain mutations.146 Hence, a positive antibody test should not be considered proof of immunity, especially with uncertainty about quantifying protection from natural or vaccine-induced immunity. This raises doubts about the reliability of commercially available immunity passports, given reduced protection against dominant variants in many countries.147,148
Contact tracing, population testing, and strategies adopted to scale up
Contact tracing
Testing and contact tracing are critical strategies for curbing the initial spread of infections within a country. Identifying and isolating infected individuals and their secondary contacts and enforcing quarantine measures for those exposed effectively halt further virus transmission. Effective large-scale contact tracing programs, particularly in East Asia, have been instrumental in controlling SARS-CoV-2 transmission. These regions’ previous experience with the 2003 SARS outbreak enabled them to deploy robust tracking mechanisms. These programs successfully identified and isolated thousands of individuals connected to an outbreak by utilizing various data sources, including patient interviews and records such as medical documents, mobile phone data, and credit card transactions. For instance, in Seoul’s Itaewon district, they traced contacts across multiple transmission cycles from the initial outbreak, as illustrated in Figure 5e.149 Its effectiveness relies on promptly identifying contacts, which is challenging because SARS-CoV-2 carriers can become infectious shortly after exposure, often before displaying symptoms. Rapid testing is crucial for successful contact tracing, with individuals advised to proactively isolate while awaiting results.150 It is a demanding and time-consuming process, particularly challenging during active viral spread. As case numbers rise, the exponential growth of secondary contacts overwhelms the identification, testing, and isolation efforts. This delay reduces the effectiveness of contact tracing. Digital solutions like mobile apps offer automation but necessitate widespread use. Beyond a certain point, when the caseload surpasses a country’s tracing capacity, contacting secondary cases becomes too late to significantly impact viral transmission.149,151
Population testing
Large-scale testing is required to mitigate the community transmission of this infection. Efforts to control SARS-CoV-2 have faced challenges as community transmission persists in many countries. This has led to a shift from contact tracing to large-scale population testing to identify asymptomatic and mildly symptomatic individuals unknowingly spreading the virus. In a pioneering move, Slovakia tested its entire population in October 2020 using rapid antigen tests, followed by isolation recommendations for positive cases and their close contacts. While this extensive effort significantly reduced infections, its impact varied, with more substantial effects in regions with high viral prevalence and limited impact in lower prevalence areas (Fig. 5c, d).152–154
The reproduction number represents the average number of secondary infections caused by one primary infected individual within a susceptible population. The calculation incorporates several factors: the virus’s transmission characteristics, the duration of infection, its potency, and the level of contact among individuals. Influences on contact level include population density, geographical location, mobility, and interventions such as social restrictions. They indicate the cumulative impact of viral spread or decline over time during a pandemic. An R-value above 1 signals an increase in viral transmission, while an R-value below 1 suggests declining transmission, indicating a decline. However, R may not fully capture the heterogeneous nature of viral spread, particularly in cases like SARS-CoV-2, where a disproportionate number of infections are driven by ‘superspreader’ events. This aspect highlights the heightened variability of SARS-CoV-2, especially in low-prevalence scenarios, setting its transmission dynamics apart from those of other pathogens, such as influenza viruses.155,156
The test positivity rate reflects the percentage of positive test results and serves as a measure of testing adequacy about viral prevalence. A low rate indicates both low viral prevalence and an effective testing system. Conversely, a high rate suggests elevated viral prevalence or biased testing toward symptomatic individuals, potentially missing many infections. An increasing rate signals rapid viral transmission. When combined with metrics like the reproductive number (R), it guides public health actions. For example, the WHO advises maintaining a rate below 5% for two weeks before altering public health measures.152 We have analyzed publically available data (https://ourworldindata.org/coronavirus ) and presented the positive rate of COVID-19 diagnosis concerning several continents (Fig. 7b).
Sensitivity in a test represents the portion of individuals correctly identified as having the condition. A highly sensitive test minimizes false negatives by effectively detecting infected individuals. Sensitivity, usually determined under controlled conditions, relies solely on test performance. In real-world scenarios, factors like sampling and processing errors can reduce sensitivity. For instance, inadequate swabbing is a significant factor, leading some approaches to employ dual testing with multiple sample types, such as nasopharyngeal and sputum or throat samples, to enhance accuracy.157 However, specificity in a test is its capacity to accurately label uninfected individuals as non-infected. Tests with high specificity minimize false positives, preventing erroneous infection diagnoses in healthy individuals. Low specificity leads to numerous false positives, causing unnecessary quarantine and treatment, which is particularly problematic in large-scale testing initiatives.79,133,134,138,152,158–160
Strategies adopted to scale up the testing
Population-scale testing commonly utilizes RT-qPCR, conducted in centralized high-throughput labs by trained personnel with automated equipment. While these labs ensure reliable results due to rigorous oversight, sample transportation to these facilities can prolong testing times to several days. Pooling samples involves testing multiple individuals simultaneously. In Qingdao, China, seven million people were tested in 3 days by combining ten samples into one test.161,162 If the pooled test is negative, all individuals are considered negative. Only if it is positive are individual samples tested separately. Another method involves overlapping pools to uniquely identify samples without extra testing. Pooling conserves reagents and increases testing capacity. However, it is less effective with high positivity rates, leading to more individual tests. Non-random pooling within households or groups can help, but pooling may introduce reporting delays and reduce sensitivity in large pools due to sample dilution.163,164 On-site, self-testing, or POC tests are performed on-site, like in clinics, workplaces, or homes. They commonly employ antigen-based lateral flow assays, are portable, need no special training or equipment, and can be widely distributed. Decentralization enhances testing access frequency and reduces healthcare worker exposure. It is an attractive option for expanding testing and includes various technologies like molecular, antigen-based, and serological approaches.165,166 In November 2020, Liverpool, UK, initiated a pilot scheme intending to screen around half a million people using on-site antigen tests. Regardless of symptoms, this program provided routine testing for all residents, aiming for widespread coverage and reduced viral transmission. While it identified over a third of infected but mildly or asymptomatic individuals, the antigen tests’ sensitivity was notably lower than in the initial validation studies, missing nearly a third of infectious cases.159,160,167,168
Emerging horizons of COVID-19 diagnosis
To address the limitations of existing detection methods for SARS-CoV-2, we propose several innovative point-of-care (POC) technologies. Drawing inspiration from established techniques used in detecting other coronaviruses, such as SARS-CoV and MERS-CoV, our approach incorporates various advanced technologies. These include the enhancement of PCR sensitivity through functionalized nanostructures, the integration of aptamers with quantum dots (QDs), the use of semiconductor-based binding assays, the application of surface plasmon resonance-based assays, the development of paper-based assay platforms, the adoption of piezoelectric immunosensors, and the advancement of electrochemical sensors. Notably, many of these technologies are scalable and suitable for large-scale testing efforts, which is crucial for identifying asymptomatic carriers and, thereby, helping to prevent further spread of COVID-19.169 In the following sections, we will explain these pivotal methods in more detail.
Lateral flow tests (LFA)
LFAs are a promising technology for swift, accurate, and cost-effective detection. LFAs offer a crucial advantage by eliminating the need for specialized equipment in qualitative tests, making them valuable for POC diagnostics. These assays comprise four essential components: the sample pad for receiving the test sample, the conjugation pad containing specific antibodies or antigens linked to labels, the membrane utilizing capillary forces to guide the sample solution to the test and control lines, and the absorption pad for sample collection. LFAs operate on a sandwich immunoassay principle. The test sample interacts with the conjugation pad, where anti-SARS-CoV-2 antibodies bind to conjugated antigens, forming complexes. These complexes advance to the test line, generating distinct signals, often in the form of colors, based on labels such as colloidal gold or carbon. LFAs primarily detect pathogen-specific antibodies, with clinical studies demonstrating an 82% sensitivity for both IgM and IgG, potentially improved by using innovative nanoparticles.153,170 Numerous LFAs are either in the developmental stages or already accessible for SARS-CoV-2 detection, primarily focusing on detecting IgM and IgG antibodies. However, these tests may yield false negatives during the early phases of infection. Although molecular tests like RT-LAMP have been integrated into LFAs for MERS-CoV, their sensitivity has proven inadequate. An innovative assay designed for Escherichia coli detection employed a hydrophilic, porous platform with photoluminescence-quenching capabilities, enabling highly sensitive detection of various targets. The key challenges associated with LFAs pertain to timing and sensitivity. To surmount these challenges, a promising avenue involves the creation of LFAs capable of directly identifying SARS-CoV-2. This can be achieved by incorporating signal amplification strategies, including plasmonic nanoparticles, carbon nanomaterials, organic compounds, and dual sensitizers, to detect even minimal SARS-CoV-2 concentrations during the early stages of infection.171–173
Paper-based devices
These devices provide a practical solution to the intricate sample preparation challenges linked to COVID-19 molecular detection tests. They seamlessly integrate various functional components with molecular amplification technologies like PCR or LAMP, enabling precise pathogen quantification. These user-friendly, portable, and efficient devices are easy to store and transport while ensuring rapid, sensitive, and accurate pathogen identification. Using a foldable, origami-like approach, they streamline sample preparation, encompassing extraction, purification, elution, amplification, and detection. Paper-based devices have demonstrated their effectiveness in detecting malaria species, providing results in under 50 min with a superior 98% sensitivity compared to commercial immunodiagnostic tests. These innovations eliminate the need for specialized laboratory equipment and infrastructure and find extended utility in diagnosing various infectious pathogens, including human papillomavirus, Zika virus, human immunodeficiency virus (HIV), and rotavirus.174,175 This technology is proposed for detecting SARS-CoV-2 in wastewater, allowing for the prediction of COVID-19 spread. Fecal and urine samples from infected individuals may introduce live virus into wastewater. Analyzing wastewater and sewage networks can help identify suspected COVID-19 cases locally, enabling measures to curb the virus’s spread. However, this analysis must be rapid, and the detection technology portable, swift, and accurate, especially for low SARS-CoV-2 concentrations. Therefore, paper-based devices are being considered for wastewater analysis, with potential challenges stemming from the complex wastewater matrix already addressed.176–178 Recently, a glycol-nanoparticle platform has identified N-acetyl neuraminic acid as a binder to the SARS-CoV-2 spike glycoprotein. Optimized nanoparticle size and coating enabled selective detection of the spike protein over SARS-CoV-1 using lateral flow assays. The paper-based system, tested with virus-like particles and pseudotyped lentivirus, demonstrated detection within 30 m, showing promise as a rapid, low-cost diagnostic tool for COVID-19.179–180
Microfluidic devices
Microfluidics in pathogen detection, including SARS-CoV-2, offer key advantages: portability, POC capability, improved surface-to-volume ratios, compatibility with small sample volumes, and efficient heat and mass transfer. These attributes enable rapid, precise, and cost-effective detection. Stability in varying conditions, user-friendliness, and specific results are crucial. Microfluidic devices with micrometer-sized channels and chambers facilitate efficient sample preparation, including high-resolution separations. They have successfully detected various biomarkers and viruses like rotaviruses, influenza, HIV, HBV, Zika, and SARS.182–184 These devices, combined with PCR and isothermal methods, allow for the simultaneous detection of multiple targets, which is crucial for diseases like SARS-CoV-2 with symptoms resembling other viral pneumonia. In HIV detection, microfluidic devices with nucleic acid probes and magnetic beads for genome purification, coupled with PCR targeting four HIV genes, significantly improved sensitivity and specificity, providing results in just 95 min. These versatile devices, already successful in detecting various viruses, have the potential for SARS-CoV-2 detection.185
Piezoelectric technology
The piezoelectric method utilizes electro-mechanical tools like quartz crystal microbalances and micro-cantilevers for virus detection.179–181 These devices comprise a mass-sensitive substrate and a piezoelectric crystal. Alterations in mass on the resonator surface, such as viral antigens or complete viruses, impact the resonant frequency. When a bio-recognition element (e.g., antibody) on the crystal surface binds to a biomolecule, it reduces the frequency due to increased mass. They are known for high sensitivity, cost-effectiveness, and specificity and are ideal for virus and bacteria detection. They are particularly valuable for POC diagnosis, including COVID-19 screenings.186
Artificial intelligence (AI)
AI can enhance COVID-19 diagnosis via chest X-rays or CT scans, particularly addressing the challenge of training experts for image analysis. AI offers rapid, cost-effective detection of SARS-CoV-2 from these scans, saving radiologists time and effort. Fueled by extensive population data, deep learning algorithms enable accurate COVID-19 diagnosis. Numerous AI applications, in development or already deployed, focused on SARS-CoV-2 diagnosis.187,188 COVID-Net, a deep convolutional neural network, leverages data from various lung conditions and SARS-CoV-2-related factors to diagnose COVID-19 via chest X-rays with 92.4% accuracy. It is open-source and accessible for various facilities. CoV-Net, a 3D deep learning model, distinguishes COVID-19 from other lung diseases with 97.17% AUC, 90.19% sensitivity, and 95.76% specificity. Another proposal involves diagnosing COVID-19 using smartphone sensors, offering a cost-effective surveillance solution. AI, coupled with deep learning, proves suitable for identifying SARS-CoV-2-related chest CT and X-ray abnormalities, given its growing implementation and track record in efficient decision-making.189–192
Conclusions
This review has explored advancements in COVID-19 diagnostics, yet several limitations must be addressed to enhance responses to novel coronaviruses and related infectious diseases. Firstly, there is an over-reliance on technological innovation with insufficient focus on equitable access to diagnostics, particularly in low- and middle-income countries. The global disparity in diagnostic capacity, exacerbated by financial, logistical, and infrastructural constraints, hinders an effective and timely response to pandemics. Without robust mechanisms for global distribution and accessibility, even the most advanced diagnostic tools will have limited impact. Additionally, the rush to deploy diagnostics during the COVID-19 pandemic revealed gaps in regulatory oversight and standardization. Although emergency-use authorizations enabled rapid deployment, the insufficient validation of these assays resulted in variable accuracy, undermining public confidence in testing. Future health policies must balance speed with rigorous evaluation to ensure quality and reliability during pandemics. There is an urgent need for governments and global health bodies to adopt a comprehensive, systemic approach to pandemic preparedness. This includes enhancing diagnostic accuracy and integrating these diagnostics within broader public health strategies. Key measures should include real-time genomic surveillance for emerging pathogens and variants, rapid adaptation of diagnostics and vaccines, and the maintenance of robust surveillance systems that detect outbreaks early. Besides technological advancements, health policies should emphasize non-pharmaceutical interventions such as social distancing, mask usage, and sanitation protocols, particularly in the initial phases of an outbreak when vaccines and treatments may not be available. Vaccination campaigns should be coordinated with diagnostic efforts to track efficacy and variant evolution in real time.
The rapid development and deployment of molecular tests, particularly RT-PCR, became the gold standard for detecting SARS-CoV-2, providing accurate and timely results. Additionally, antigen tests offered faster, more accessible options, though their sensitivity varied depending on the infection phase. Serological tests also played a role by identifying past exposure and aiding in understanding population-level immunity. Despite these achievements, challenges such as ensuring equitable access to testing, addressing false negatives or positives, and adapting diagnostics for emerging variants persist. As the global response continues, refining diagnostic technologies and strategies will be pivotal in managing COVID-19 and potential future coronavirus outbreaks. Integrating AI and machine learning can enhance applicability and accuracy. Investment in surveillance systems and rapid testing infrastructure is also crucial for early detection. These advancements promise enhanced capabilities for managing COVID-19 and provide a blueprint for responding to future pandemics with flexibility and precision.
In conclusion, improving diagnostic approaches for future pandemics requires more than technological breakthroughs; it necessitates a global commitment to equitable access, regulatory rigor, and comprehensive preparedness strategies. By incorporating accurate diagnostics, vaccination, and public health measures, future pandemics can be met with coordinated, effective responses that protect global health and mitigate the devastating impacts seen during COVID-19. As we continue to navigate the evolving landscape of COVID-19 and prepare for future pandemics, the lessons learned and the innovative diagnostic strategies explored in this report offer hope and readiness, showing the imperative of preparedness, collaboration, and adaptability in safeguarding global health.
Declarations
Acknowledgement
Not applicable.
Data sharing statement
Data were obtained from the analysis of publicly available datasets
Funding
This work is jointly supported by the Department of Science and Technology (NanoMission: DST/NM/NT/2018/105(G); SERB: EMR/2017/000992) and Focused Basic Research (FBR), HCT, CSIR, Govt. of India.
Conflict of interest
One of the authors, Mrinal K Ghosh, has been an editorial board member of Nature Cell and Science since June 2023. The authors have no other conflict of interest to note.
Authors’ contributions
Conceived the idea and review structure (SK and MKG), Wrote the manuscript (SK, DC, and MKG), Revised and edited the manuscript (SK, PG, MB, and MKG). All authors have read, agreed, and approved the final draft of the manuscript.

 Author information
Author information