Introduction
From the mid-2000s to the early 2010s, a series of nuclease-based genome-editing tools and technologies were developed, and several genetically modified animals in different species were produced using meganucleases,1 zinc-finger nucleases (ZFNs),2–4 or transcription activator-like effector nucleases (TALENs).5–7 In late 2013, genome editing technology was significantly accelerated with the appearance of clustered regularly-interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), which is more convenient and simpler than other methods such as ZFNs and TALENs.8,9 The CRISPR/Cas9 system is based on two factors: guide RNA (gRNA), which binds to a specific chromosomal DNA site, and the Cas9 endonuclease, which cleaves double-stranded (ds) DNA three bases upstream of the protospacer adjacent motif, under the guidance of the gRNA.8,9 After the cleavage of dsDNA, double-strand breaks are introduced at the target site. The resulting double-strand breaks are repaired via a non-homologous end-joining (NHEJ)-based process, which frequently leads to nucleotide insertions or deletions (indels). The appearance of indels at a target site often causes the formation of premature termination (stop) codons, leading to the failure of protein expression due to nonsense-mediated messenger RNA (mRNA) decay, a translation-dependent surveillance mechanism in eukaryotes.10 Thus, these indels lead to the generation of a knockout (KO) phenotype in an organism. In contrast, homology-directed repair (HDR) occurs at the cleavage site in the presence of a donor DNA sequence. This event is called the HDR-mediated knock-in (KI) of the donor sequence. Notably, the efficiency of HDR-mediated KI of the donor sequence is generally found to be lower than that of NHEJ-mediated genome editing. Furthermore, HDR preferentially occurs in dividing cells, whereas NHEJ occurs in both dividing and non-dividing cells.11 The use of NHEJ inhibitors has been employed to elevate HDR efficiency by suppressing the NHEJ pathway in cultured cells.12,13 Unfortunately, there is no standardized method that can control the efficiency of HDR at present.
Recently, base editing (BE) and prime editing systems, which are derivatives of CRISPR/Cas9 that can introduce specific changes to DNA, have been developed.14,15 In BE, two classes of DNA base editors—cytosine BEs (CBEs) and adenine BEs—have been described. Prime editing has expanded the CRISPR-based editing toolkit to include all 12 possible transition and transversion mutations and small insertion or deletion mutations. With these techniques, manipulating maternal mitochondrial DNA (mtDNA) has attracted increased attention, especially in oocytes and early embryos. For example, double-stranded DNA-specific cytidine deaminase-derived CBEs (DdCBEs), which catalyze C•G-to-T•A conversion through a modified double-stranded DNA-specific cytidine deaminase, were first reported to be useful for manipulating mtDNA in human cells.16 Wei et al.17 tested the ability of DdCBE to edit mtDNA in human embryos. When clinically discarded human embryos with three pronuclei were injected with DdCBE mRNA, the DdCBE was found to be an effective BE for inducing point mutations in mtDNA of human embryos, and the efficiency was much higher in 8-cell embryos. This approach will enable the generation of mitochondrial disease models and derived embryonic stem cells for the functional investigation of disease-associated mutations in mtDNA.
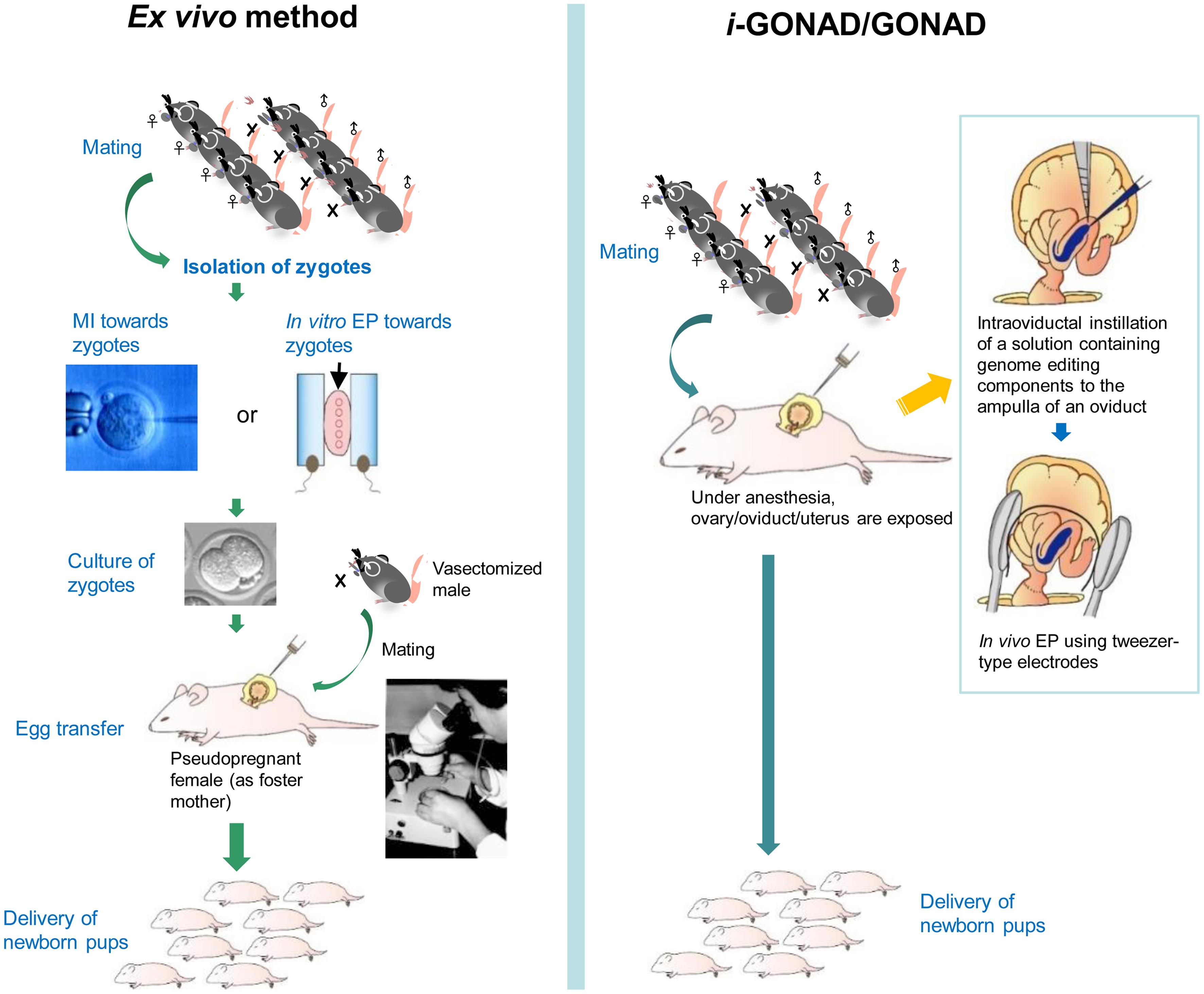
Gene editing in mammalian zygotes was first performed using pronuclear or cytoplasmic microinjection (MI) of genome editing reagents into zygotes isolated from pregnant female rodents or through in vitro fertilization (IVF).18–21 In 2014, the usefulness of in vitro electroporation (EP) for producing genome-edited rats was first reported by Kaneko et al.22 Since then, numerous genome-edited (GE) mice and rats have been successfully produced using in vitro EP.23–25
The MI and in vitro EP approaches for generating GE animals have specific properties (Fig. 1a). For example, in the case of MI, large DNA (>10 kb) can be introduced into zygotes. However, this requires a skilled person to manipulate the embryos under a microscope, making the process time-consuming. In general, it takes over 2 h to treat approximately 100 zygotes.26 In the case of in vitro EP, an expensive electroporator is required, but this method does not require a skilled person, unlike MI, and many zygotes (over 100) can be genome-edited within a short period. For example, approximately 30 zygotes can be electroporated within 15 m.26 However, it cannot incorporate large DNA such as plasmid DNA. Both systems require ex vivo handling of embryos (i.e., isolation of zygotes from a pregnant mother to perform genome editing in vitro, culture of treated embryos for a short period, and egg transfer (ET) of genome-edited embryos to the reproductive tract of a pseudopregnant female) (Fig. 1a).
MI, microinjection; EP, electroporation; ET, egg transfer.
In 2015, Takahashi et al.27 first reported a novel method called genome editing via oviductal nucleic acid delivery (GONAD) (Fig. 1b), which enables genome editing in mouse two-cell embryos in situ. In this system, a solution (1–1.5 μL) containing Cas9 mRNA, single guide RNA (sgRNA), and trypan blue (a visible marker for monitoring the injection process) was injected via the oviductal wall into the ampulla of the oviduct of a pregnant female (corresponding to the two-cell stage) using a mouthpiece-controlled glass micropipette. Immediately after injection, the entire oviduct was held using tweezer-type electrodes and electroporated using a square pulse generator (electroporator). Under appropriate electric conditions, the genome editing components present outside the embryo (floating in the oviductal lumen) are transferred through the zona pellucida (ZP) to the inside of the embryo. In 2018, Ohtsuka et al.28 improved GONAD and re-named it “improved GONAD (i-GONAD).” The major improvements were the use of late zygotes to avoid possible mosaicism, as has been frequently observed with GONAD, and the employment of ribonucleoprotein (RNP) (comprised of Cas9 protein and crRNA/tracrRNA) to induce genome editing more rapidly than when using Cas9 mRNA. As a result, embryos with indels at the target locus were obtained with an efficiency of 97%. When a solution containing RNP, single-stranded oligodeoxynucleotide (as a DNA donor), and Fast Green FCF (as a visible marker like trypan blue) was used for i-GONAD, the KI embryos were recovered with efficiencies of ∼50%. GONAD/i-GONAD does not require ex vivo handling of embryos and has been frequently employed for MI or in vitro EP-mediated production of genome-edited animals. Therefore, GONAD/i-GONAD is more convenient than previous methods based on MI or in vitro EP. For instance, it requires only four to six pregnant females for one session and finishes within 15 m for both oviducts.26 This fits the 3R principle (Replacement, Reduction, and Refinement), an international tenet for animal experimentation, aimed at reducing the number of animals used whenever possible.
As aforementioned, several gene delivery methods, including physical delivery such as MI and EP and viral-based delivery, have been employed for generating GE mammalian embryos.29 Concerning viral-based delivery, viral vectors such as lentiviral, adenoviral, retroviral, and adeno-associated viral (AAV) vectors are most widely used for therapeutic or experimental purposes.30 Among these vectors, only AAV vectors are known to infect ZP-enclosed early mammalian embryos after simple in vitro incubation in a medium containing AAV vectors for a short period (∼16 h).31–33 When screening was performed to determine which AAV serotype was appropriate for the transduction of ZP-enclosed mouse embryos, AAV serotype 6 (AAV6) was found to be most effective.32,33 Furthermore, AAV6 infected ZP-enclosed rat and bovine embryos, suggesting the utility of this vector for manipulating early embryos across species.33 Notably, intraoviductal instillation of a solution containing AAV6 carrying the gene for Neisseria meningitidis (Nme) 2 Cas9 (Nme2Cas9) (which is smaller in size than the more widely used Streptococcus pyogenes-derived SpCas9) and the gRNA expression unit into a pregnant female mouse was shown to be a useful method for obtaining GE animals, similar to GONAD/i-GONAD, but performed without in vivo EP.34 Since then, this AAV-based method for in situ acquisition of GE animals has been called “AAV-based GONAD”.29,35
However, to the best of our knowledge, large animals remain challenging for GONAD/i-GONAD-mediated gene editing. Many GE large animals, including ferrets, cats, dogs, rabbits, sheep, goats, cattle, pigs, and monkeys, have been produced using MI, in vitro EP, and somatic cell nuclear transfer (SCNT) of GE somatic cells.36–38 Typical examples regarding the production of large GE animals (ferrets, pigs, and monkeys) are shown in Table 1,39–52 as these animals are considered important experimental subjects for biomedical research and the creation of human disease models. In this review, we intended to apply GONAD/i-GONAD to large experimental animals (especially ferrets, pigs, and marmosets), as it has the potential to bypass traditional approaches for GE animal production that rely on the “ex vivo handling of embryos.”
Summary of the production of genome-edited (GE) large animals (ferrets, pigs and monkeys)
| Animals | Method for gene modification | GE Tool (mode for Gene modification) | Outcome | Target Gene | References |
|---|---|---|---|---|---|
| Ferrets | MI (Cas9 mRNA and sgRNAs) | CRISPR (indels) | Generation of F0 KO ferrets with an efficiency up to 73%.; showing a lissencephaly phenotype similar to human Doublecortin patients | Dcx, Aspm, Disc1 | Kou et al. (2015)39 |
| Ferrets | MI (mRNA) | TALENs (indels) | All 11 alive ferrets had insertions/deletions (100% efficiency); KO offspring exhibited severe microcephaly (25–40% decreases in brain weight). | Aspm | Johnson et al. (2018)40 |
| Ferrets | MI (HITI-based donor plasmid DNA) | CRISPR (KI) | The KI floxed Cre-reporter ferrets were generated with an efficiency of 22%; showing high expression of a transgene in all tissues. | Rosa26 | Yu et al. (2019)41 |
| Ferrets | MI (Cas9 mRNA and sgRNAs for KO or Cas9 mRNA, sgRNAs and oligo for KI) | CRISPR (indels, KI) | The KO ferrets had alpha-1 antitrypsin deficiency (AATD) with airflow limitation and obstruction; the KI ferrets with E342K point mutation exhibited a severe form of AATD. | SERPINA1 | He et al. (2022)42 |
| Pigs | SCNT | ZFNs (indels) | All 6 fetuses obtained lacked α-Gal epitopes. | GGTA1 | Hauschild et al. (2011)43 |
| Pigs | MI (mRNA) | TALENs (indels) | Ten of 18 live-born clones contained biallelic KO for LDLR. | LDLR | Carlson et al. (2012)44 |
| Pigs | EP (RNP) | CRISPR (indels) | EP of zygotes resulted in efficient production of MSTN KO pigs. | MSTN | Tanihara et al. (2016)45 |
| Pigs | MI (Cas9 mRNA, sgRNA and oligo) | CRISPR (KI) | Three of 5 piglets delivered exhibited pigmentary disorders with light-colored iris in eye. | Sox10 | Zhou et al. (2016)46 |
| Monkeys (rhesus and cynomolgus monkeys) | MI (plasmids) | TALEN (indels) | Male rhesus (2) and cynomolgous (1) fetuses carrying MECP2 mutations were miscarried during mid-gestation, consistent with Rett syndrome (RTT)-linked male embryonic lethality in humans. | MECP2 | Liu et al. (2014)47 |
| Monkeys (cynomolgus monkeys) | MI (Cas9 mRNA and sgRNAs) | CRISPR (indels) | Simultaneous disruption of two target genes (Ppar-γ and Rag1) in one step with no off-target mutagenesis. | Ppar-γ, Rag1 | Niu et al. (2014)48 |
| Monkeys (cynomolgus monkeys) | MI (Cas9 mRNA, sgRNA and donor plasmid) | CRISPR (KI) | KI of an Actb-p2A-mCherry insert into the Actb gene was performed in monkeys to achieve mCherry expression under the control of the Actb promoter. | Actb | Yao et al. (2018)49 |
| Monkeys (rhesus monkeys) | MI (Cas9 mRNA and gRNAs) | CRISPR (indels) | One PINK1 KO monkey reduced its food intake and showed weakness at the age of 1.5 years, suggesting neurodegeneration in its brain. | PINK1 | Yang et al. (2019)50 |
| Monkeys (cynomolgus monkeys) | MI (BE4 mRNA and gRNA) | CRISPR (BEs) | Hutchinson-Gilford progeria syndrome (HGPS) monkey model recapitulated the typical HGPS phenotypes including growth retardation, bone alterations, and vascular abnormalities. | LMNA | Wang et al. (2020)51 |
| Monkeys (cynomolgus monkeys) | MI (Cas9 nickase mRNA and truncated sgRNAs) | CRISPR (indels) | Three newborns exhibited frame-shift mutations of PINK1, but the protein levels were unaltered, suggesting no effects on protein expression in the case of use of pair truncated sgRNA/Cas9 nickase. | PINK1 | Chen et al. (2021)52 |
Possible application of GONAD/i-GONAD to ferrets
Ferrets (Mustela putorius furo) are considered one of the most valuable animal models for recapitulating human diseases, as they exhibit several similarities to humans with respect to brain function, reproductive biology, and susceptibility to various diseases such as cancer, influenza, and cystic fibrosis.53,54
According to Lindeberg,55 female ferrets reach puberty at eight to twelve months of age. The domestic female ferret is a seasonally polyestrous species. Proestrus begins in January or February, during which progressive vulvar swelling develops into an enlarged and edematous vulva, characterizing estrus. Estrus can persist for up to five months; however, once ovulation occurs, either pregnancy or pseudopregnancy ensues. Ovulation is induced by pressure on the cervix and is associated with copulation. After sufficient luteinizing hormone release, the pre-ovulatory follicles mature, and an average of 12 oocytes (5–13) per female ovulate into the ovarian bursa 30–40 h post-copulation (hpc). The gestation length is 41 days (39–42 days).55 Notably, the initiation of gonadal activity in female ferrets is entirely dependent on the light-dark cycle. For example, ferrets begin to exhibit estrus approximately three weeks after an artificial change from short days (8 h of light and 16 h of darkness) to long days (16 h of light and 8 h of darkness).55
Regarding the history of gene manipulation in ferrets, GE ferrets were first generated as cloned animals using SCNT of GE fibroblasts (gene-targeted via infection with an AAV vector) into enucleated oocytes to model cystic fibrosis, a recessively inherited genetic disease caused by mutations in the gene encoding cystic fibrosis transmembrane conductance regulator.56–58 Genome editing technology later enabled the direct generation of GE ferrets. For example, when TALEN plasmid-derived mRNAs targeting Aspm (abnormal spindle-like microcephaly-associated), the most common microcephaly gene mutated in humans, were injected into the cytoplasm of zygotes collected from the mating of ferrets, the resulting offspring exhibited severe microcephaly (with a 25–40% decrease in brain weight).59 Kou et al.39 demonstrated that co-injection of Cas9 mRNA and sgRNAs into ferret zygotes led to the production of KO ferrets for the doublecortin gene with high efficiency (up to 73%). The resulting F0 animals carrying doublecortin mutations exhibited a lissencephaly phenotype similar to that observed in human Doublecortin patients. Furthermore, GE ferrets with a reporter allele conditionally knocked into the Rosa26 locus were generated through MI of homology-independent targeted insertion system-based donor plasmid DNA into ferret zygotes.41 This system enabled the creation of transgenic (Tg) animals (referred to as “ROSA26-CAG-LoxPtdTomatoStopLoxPEGFP (ROSA-TG) Cre-reporter”) expressing a dual-fluorescent Cre-reporter system flanked by PhiC31 and Bxb1 integrase attP sites. The encoded tdTomato transgene was highly expressed in all tissues evaluated. Targeted integration and germ-line transmission were verified by molecular biological analysis. The function of the resulting reporter animals (showing the conversion from red to green fluorescence expression) was confirmed in primary cells following Cre expression. These models will be useful for biomedical research involving lineage tracing, evaluation of stem cell therapy, and transgenesis in ferret models of human disease. To our knowledge, ex vivo handling of ferret zygotes is currently the main approach for generating GE ferret individuals, and there has been no in vivo gene delivery (such as i-GONAD or in vitro EP) used in ferrets for this purpose.
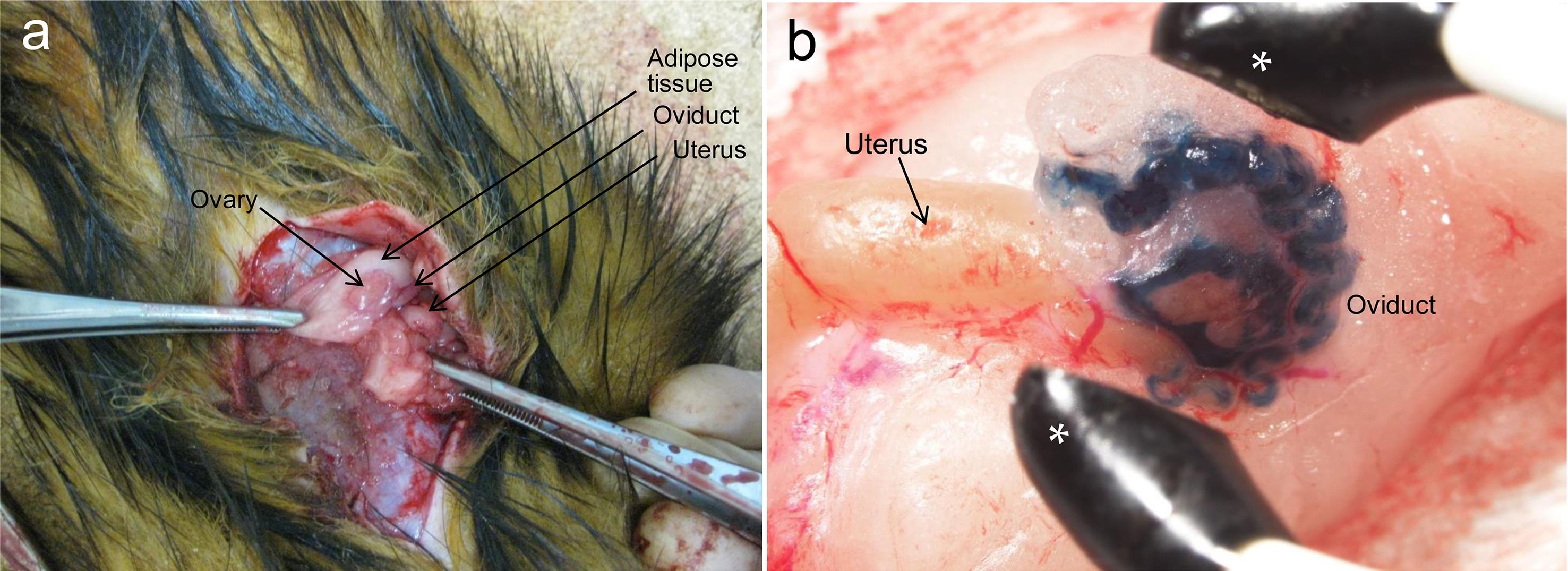
The following procedure is envisaged when i-GONAD is applied to ferrets: Ferret zygotes can generally be recovered from the oviducts at day 2 (∼45 hpc) after the first mating.59 Under anesthesia, a 3–4 cm incision is made along the midline of the abdomen of a pregnant female (on day 2; ∼45 hpc after the first mating) to expose the ovaries, oviducts, and uterus (Fig. 2a). Approximately 2 µL of a solution can be injected under observation with a dissecting microscope through a puncture in the oviductal wall using a fine glass pipette. Notably, the instillation of the solution throughout the oviduct would be visible due to the co-injection of a marker dye (Fig. 2b). It may also be possible to perform in vivo EP on the injected oviduct, as the size of the tweezer-type electrodes is sufficient to cover the entire oviduct, as shown by the asterisks in Fig. 2b. Unfortunately, we have not yet tested whether in vivo zygotes can be successfully genome-edited using this system. After in vivo EP and subsequent closure of the muscle layer and skin, the ferrets would be returned to their cages.
When the abdominal portion of an anesthetized female ferret is opened, reproductive organs such as ovary, oviduct and uterus are visible (a). When dye-containing solution is injected through oviductal wall into an oviductal lumen of a female ferret under a dissecting microscope using a breath-controlled glass micropipette, the introduced solution is visible throughout an entire oviduct (b). Asterisks, tweezer-type electrodes used for i-GONAD in mice.
Possible application of GONAD/i-GONAD to pigs
Domestic pigs are considered valuable experimental animals for biomedical applications because they exhibit physiological and anatomical similarities to humans and can accurately recapitulate the symptoms of human diseases.60 They are reproductively active throughout the year, can deliver multiple piglets in one pregnancy (averaging 14 piglets), and have a relatively short gestation period (114 days).61
Regarding gene manipulation in pigs, the generation of genetically modified (GM) pigs was first carried out via SCNT of GE somatic cells or those carrying transgenes.62,63 For example, GE pigs lacking both alleles of the α-1,3-galactosyltransferase gene were first produced through SCNT of bi-allelic KO cells using ZFNs.43 Since then, most experiments have focused on using SCNT-based production of GM piglets. However, manipulation of porcine somatic cells for HDR-based KI, the introduction of subtle changes (gene correction or point mutation), or indels (for KO) has resulted in poor efficiencies.64 Furthermore, the efficiency of SCNT-based production of GE piglets has been extremely low (only 1.0–5.0%).65 Direct MI of genome-editing components into porcine zygotes is an alternative method to produce GE piglets.63 This approach appears to be simpler and more efficient than SCNT-based gene manipulation because genome editing can be easily induced by the one-step introduction of Cas9 mRNA (or protein), gRNA, and single-stranded oligodeoxynucleotide in the case of HDR-based KI, into porcine zygotes. This method has resulted in the generation of bi-allelic KO piglets with an efficiency of up to 100%,66 although some experiments suggest that a high degree of mosaicism is often associated with this technique.63 Alternatively, in vitro EP has been recognized as a useful tool for the production of GE piglets.63 For example, Tanihara et al.45 performed in vitro EP on IVF-derived porcine embryos (13 h after IVF) in the presence of RNP and found successful genome editing in 67% (10/15) of the treated embryos (blastocysts). They further investigated whether mutant pigs could be generated using the in vitro EP method. When the in vitro EP-treated zygotes were transferred into the oviducts of two recipient gilts approximately 12 h after EP, one of the recipients became pregnant and gave birth to 10 piglets 111 days after zygote transfer. One (10%) of the piglets died soon after birth. However, nine of the ten piglets carried the KO mutations in the myostatin gene. The authors named this process “gene editing by electroporation of Cas9 protein (GEEP).”
Interestingly, a new type of genomic safe harbor (GSH) locus, similar to the Rosa26 locus, was recently found in the porcine genome.67 A GSH is a locus into which a transgene can be knocked in, and the integrated transgene can be expressed without altering the expression of other genes. Chen et al.67 performed KI of an EGFP-containing donor DNA fragment into the 3rd exon of the ubiquitously transcribed tetratricopeptide repeat-containing Y-linked gene (UTY) located on the Y chromosome using CRISPR/Cas9-mediated homology arm-mediated end joining. The unbiased expression of the integrated transgene was confirmed by molecular biological analysis. This finding indicates that the UTY locus serves as a GSH site for gene editing in the pig genome. More interestingly, this site located on the Y chromosome can be utilized for sex-biased pig breeding and developing biomedical models.
The fact that the genomes of porcine zygotes can be successfully manipulated through in vitro EP-based genome editing45 indicates that in vivo porcine zygotes can be targets for GONAD/i-GONAD-based genome editing. Notably, in vivo zygotes were surgically retrieved from naturally mating gilts. According to Wu et al.,68 female pigs were monitored for estrus twice daily by observing their responses to a mature boar, reddening of the vulva, and vaginal mucus secretion. The gilts were mated immediately with a mature boar after estrus detection. After 24 h, the urogenital tract was excised under general anesthesia. The number of zygotes in the oviduct was estimated by examining the ovaries and counting ruptured follicles. In this context, GONAD/i-GONAD may be possible approximately 24 h after successful male mating. Hormone administration can also induce estrus in female pigs. For example, estrus and ovulation were induced by intramuscular injection of 400 I.U. equine chorionic gonadotropin and 800 I.U. human chorionic gonadotropin, followed 72 h later by intramuscular injection of 750 I.U. human chorionic gonadotropin.68 Estrus was checked twice daily by exposing the sows to a mature boar (nose-to-nose contact) and applying manual back pressure.69 Females exhibiting a standing estrous reflex were used as recipients for mating with males and subsequent embryo isolation.
To perform GONAD/i-GONAD in pigs, two methods, surgical and non-surgical, are possible. According to Na et al.,70 surgical techniques have always been limited due to the complicated nature of the surgeries and the need to learn the procedures. On the other hand, the laparoscopic approach (a minimally invasive surgical procedure) can be recommended because it does not require extensive surgical time and is relatively non-invasive, as already shown in human surgeries. Its technique is already available for pigs.71 According to Wieczorek et al.,71 transfer of embryos by the infundibulum is not recommended due to handling problems and very low efficiency, as transfer of vitrified embryos resulted in poor outcomes. They preferred this method for transferring embryos to the fallopian tube, which can be used regardless of the developmental stage of the embryo. This approach has been employed in many other laboratories as well.72
When GONAD/i-GONAD is applied to pigs, the following procedure is envisaged: After mid-ventral laparotomy of an anesthetized pig, a solution containing genome-editing reagents is surgically transferred to the tip of the uterine horn (15–20 cm from the uterotubal junction; ampullary-isthmic junction of the oviduct) using an embryo transfer catheter inserted through the uterine wall of the recipient gilt, which was previously punctured with a blunt needle. In the case of GONAD/i-GONAD using mice and rats, in vivo EP has generally been performed using tweezer-type electrodes directed toward the entire oviduct after oviductal instillation. However, it may be difficult to apply in vivo EP to porcine oviducts because the oviduct itself is linear (approximately 10 cm in length),73 and there is no clear ampulla (swelling of the oviduct), as seen in mice and rats. To overcome this issue, AAV-based GONAD may be preferable, as genome editing at a target locus can be induced without using in vivo EP, as previously described by Yoon et al.32 in mice. In this case, a large amount of AAV solution will be required to achieve effective transduction of zona pellucida-enclosed in vivo zygotes. For example, during ET in pigs, intra-oviductal instillation of a smaller volume (1.6 mL) has shown the most efficient delivery rate.70 Injection of a solution greater than 1.6 mL may result in flowback to the vagina.
Possible application of GONAD/i-GONAD to marmosets
As shown in Table 1, cynomolgus and rhesus monkeys, both Old World monkeys, have been extensively used to create GE animals as non-human primates (NHPs). The Common marmoset (Callithrix jacchus) is a more primitive NHP, belonging to a species of New World monkeys. It is widely used in fundamental biology, pharmacology, and toxicology studies. Common marmosets are small and easy to handle, with rapid growth and sexual maturation (1.52 years), which are helpful for expediting breeding programs. The ovarian cycle of adult female marmosets is similar to that of humans, with a 28-day cycle. They have a lifespan of 12–15 years, and sexual maturation occurs within 1.5 years. Common marmosets typically have litter sizes of 1–3, with relatively short gestation intervals (145–148 days).74 These properties make them suitable for the production of genetically modified animals.75 In the following section, we focus on the historical background of genetic manipulation of marmosets and the possible application of GONAD/i-GONAD in these animals toward the creation of GE marmosets.
In 2009, Sasaki and her colleague76 first produced Tg marmosets carrying the green fluorescent protein gene. Notably, these monkeys passed the transgenes on to the next generation, suggesting the possibility of establishing NHP models of human diseases. Since then, there have been several reports of generating GE NHPs in the genus Macaca, such as rhesus and cynomolgus monkeys, using genome editing technologies such as TALENs and CRISPR/Cas9.77,78 The genes knocked out include methyl CpG binding protein 2,47 peroxisome proliferator-activated receptor gamma,48 X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism (DAX1 mutation),79 dystrophin gene,80 beta-actin,49 octamer binding factor 4,81 and polycystic kidney disease 1.82 Both Tg and KO marmosets have been successfully generated by virus infection83,84 and genome editing methods.85–87
Generally, the following steps are needed for conventional methods to generate GE marmosets: (1) collection of oocytes from female ovaries (called “donors”), (2) in vitro maturation of oocytes and IVF with sperm from males (notably, the current IVF rate in marmosets is around 50%, and unlike in mice, marmoset superovulation has not been established), (3) MI of genome editing reagents into embryos and culture in vitro for several days, and (4) ET of MI-treated embryos into the uteri of another set of females (called “recipient females”). This process requires a long period of in vitro culture and involves many donors and recipients. Additionally, conventional methods require parous females as recipients, which can be difficult to identify when the number of females is limited. Abe et al.87 reported a novel convenient method for creating CRISPR-based GM marmosets using autologous embryo transfer (AET), which is routinely employed in humans as an assisted reproductive technology to treat infertility. It should be noted that AET is not frequently used in experimental animals. AET uses a female marmoset as the recipient from which the embryos are provided, thus fitting the 3R principles. In contrast, the manipulation of isolated zygotes (∼2 per female), i.e., cytoplasmic MI of CRISPR components, must be performed within 30 m. To elaborate, at the zygote stage, the oviducts and ovaries of female marmosets were exteriorized by midline laparotomy and placed in a 60-mm culture dish. Notably, the infundibulum of the marmoset oviduct is not covered by the ovarian bursa and is usually open. To isolate the zygotes, a 27-gauge winged needle was inserted into the isthmus of the oviducts of a naturally mating female. The pronuclear stage zygotes were recovered from the oviducts by flushing. Embryos were immediately cytoplasmically injected with Cas9 protein and gRNAs targeting the fragile X mental retardation 1 gene, which encodes the fragile X mental retardation protein. Of the 29 zygotes injected with CRISPR reagents, 27 were transferred into the oviducts of 14 females. Of the 14 AET-treated mothers, eight successfully delivered 10 pups, six of which were identified as having indels.
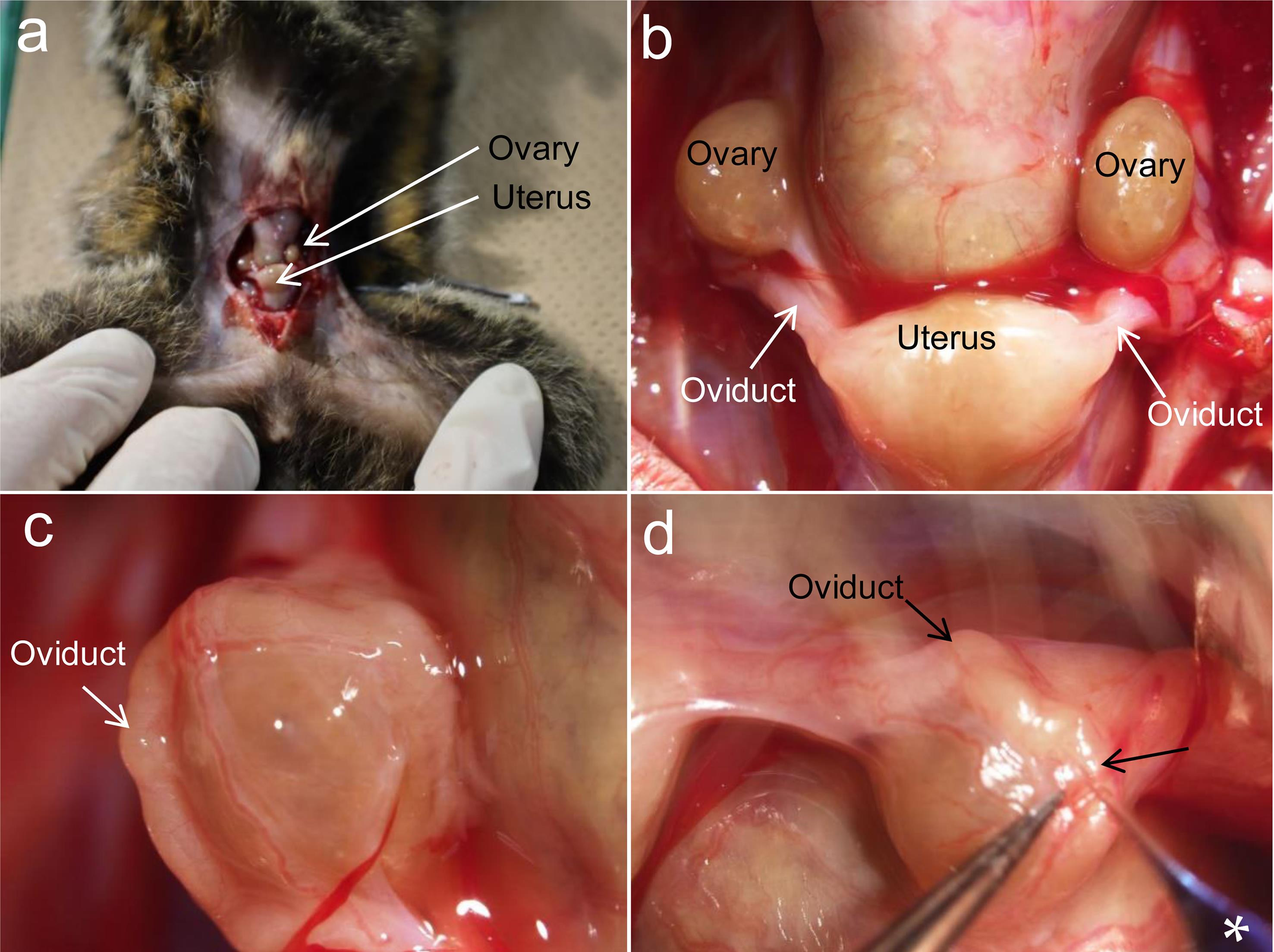
As aforementioned, GE monkeys, including marmosets, have been produced through viral infection or MI of editing reagents into the cytoplasm of zygotes under ex vivo conditions. To the best of our knowledge, EP-based gene modification of early embryos has not been reported for the purpose of producing GE monkeys. This prompted us to employ AAV-based GONAD to produce GE marmosets through in vivo manipulation. In this case, it may be difficult to introduce a solution via the oviductal wall, such as GONAD/i-GONAD, because we failed to puncture the oviductal wall with a glass pipette (Fig. 3). Notably, Abe et al.87 successfully transferred ex vivo-treated marmoset embryos via the infundibulum. It is likely that the AAV vector-containing solution can be injected via the infundibulum of a pregnant female marmoset. However, there is a major challenge in the large-scale manufacturing of AAVs, which is costly and labor-intensive.
When the abdominal portion of an anesthetized female marmoset is opened, reproductive organs such as ovary and uterus are visible (a). Upon magnification, oviduct becomes visible in tight association with uterus (b, c). When a dye-containing solution is attempted to inject through oviductal wall into an oviductal lumen of a female marmoset under a dissecting microscope using a breath-controlled glass micropipette, it failed, due to that it was too hard to penetrate the oviductal wall by a glass micropipette (d). Arrow in (d) indicates the site at which the micropipette was tried to insert.
When AAV-based GONAD is applied to generate GE marmosets, the following procedure based on a study by Abe et al.87 may be preferable. For instance, to reset the estrous cycle of female marmosets, a prostaglandin analog should be administered on day 0. On day 1, the concentration of progesterone (P4) in the blood should be confirmed to be below 10 ng/mL using a competitive enzyme immunoassay. On day 6, females should be mated with mature male marmosets. From day 8, successful mating should be confirmed by checking the vaginas of the females daily for the presence of sperm. Subsequently, the blood levels of estradiol (E2) and P4 should be measured daily using competitive enzyme immunoassays. The presence of zygotes in the oviduct can be inferred when the level of P4 increases and that of E2 decreases compared to the previous day. Both the oviducts and ovaries should be exteriorized by midline laparotomy, and a solution containing AAV in a glass pipette should be inserted via the infundibulum and injected.
Limitations of GONAD/i-GONAD
GONAD/i-GONAD employs in vivo EP to enhance gene delivery. However, in contrast to MI, which enables the introduction of large-sized DNA, the EP-based introduction of long DNA fragments into mammalian zygotes within the oviduct is currently challenging. Quadros et al.88 and Miura et al.89 succeeded in introducing a long single-stranded DNA fragment (approximately 1 kb in size) using GONAD/i-GONAD. At present, the targeted introduction of a long single-stranded DNA (lssDNA) donor of more than 1 kb remains difficult when GONAD/i-GONAD is used. Notably, Inoue et al.90 optimized the phospho-PCR method to obtain highly pure lssDNA suitable for generating KI mice via genome editing. They used two exonucleases (Lambda exonuclease and Exonuclease III) to obtain lssDNA (∼2.7 kb) without the need for laborious gel extraction steps: initially, dsDNAs were amplified using a pair of 5′-phosphorylated primers and normal primers. The resulting products, where one strand was phosphorylated, were then treated with lambda exonuclease, an enzyme that preferentially digests the phosphorylated strand, to yield ssDNA. Treatment with Exonuclease III then degraded the remaining dsDNA. To confirm the functionality of the obtained lssDNA, Inoue et al.90 microinjected these donor DNAs along with CRISPR/Cas9 components into mouse zygotes, resulting in viable KI strains. Furthermore, they demonstrated that in vitro EP, in the presence of highly concentrated lssDNA donors, resulted in the generation of KI embryos with efficiencies of 10–14%. This encouraged us to perform i-GONAD using an ssDNA donor prepared by the phospho-PCR method to generate mouse strains with longer KI inserts in their genome.
The use of CRISPR/Cas9 is associated with the issue of off-target effects, which can potentially diminish the efficacy of CRISPR-Cas9. However, recent progress has demonstrated that this possibility can be substantially reduced. For example, various methods for modifying genomic RNA (such as adjusting GC content, sgRNA length, truncated sgRNA, and chemical modifications) have been developed.91 The development of improved Cas variants with reduced non-specific binding to non-targeted DNA has also been extensively explored. For example, SpCas9 mutants, such as enhanced SpCas9 and SpCas9-HF1 (high-fidelity variant-1), appear to address this concern.92 Potential immune responses are also a major concern, especially when AAV-based GONAD is applied to larger animals. According to Ronzitti et al.,93 the current preclinical evidence shows that recombinant AAV vectors can trigger both innate and adaptive immune responses. Special attention is required when performing gene delivery experiments using AAV-based GONAD.
At present, due to the difficulty in accessing the oviduct via surgery, the need to inject large amounts of solution into the oviduct, and the challenges of in vivo EP using tweezer-type electrodes, the application of GONAD/i-GONAD to pigs and marmosets appears unrealistic. However, we have successfully injected the solution throughout the entire oviduct of a ferret after oviductal puncture using a glass micropipette, similar to the i-GONAD technique used in mice (Fig. 2). This success has encouraged us to perform i-GONAD to generate GE ferrets.
Conclusions
Advances in genome-editing technology have made it possible to generate GE animals in a relatively short time through the direct introduction of genome-editing reagents into zygotes. Consequently, numerous genome-edited animal models, including mice and rats, have been developed over the past decade. However, in the case of large animals, such as ferrets, pigs, and marmosets, acquiring large GE animals is more challenging compared to small-sized mice and rats due to limitations in the number of animals bred, the number of embryos obtained, the need for ex vivo handling of embryos, and the EP-based gene delivery in some species. The most beneficial aspect of using large animals is that they can recapitulate human properties due to their close similarity to humans, providing an opportunity to develop human disease models. In this review, we discuss the potential of using GONAD/i-GONAD to conveniently generate large GE animals. As mentioned previously, the application of GONAD/i-GONAD in pigs and marmosets is currently unrealistic for several reasons. However, the present results, in which the intra-oviductal injection of a solution was successful in a female ferret, encourage us to perform GONAD/i-GONAD to generate GE ferrets.
Declarations
Acknowledgement
We thank Kazusa Inada for her support with the in-house drawings shown in Figure 1. The language of the manuscript was partially edited by an external language editing service provider.
Funding
This study was partially supported by a grant (No. 23K19334 for SW, No. 21K05890 for ST, and No. 23H02404 for SN) from the Ministry of Education, Science, Sports, and Culture, Japan.
Conflict of interest
None.
Authors’ contributions
Study concept and design (SW, MS), data acquisition (SW, ST), drafting of the manuscript (MS), and critical revision of the manuscript (KM, SN).

 Author information
Author information