Introduction
Cancer drug development in the modern era focuses on identifying inhibitors of gene products directly involved in the pathogenesis of cancers. These new molecularly targeted drugs are more selective and specific for cancer types that express the variant gene product. Therapy-relevant targets have traditionally been identified through molecular testing methodologies such as polymerase chain reaction, fluorescent in situ hybridization (FISH), and massive parallel sequencing or next-generation sequencing (NGS) methods. The latter can serve as screening or definitive tests, highlighting the presence of targeted gene variants and helping in selecting the most appropriate agents for clinical use or further research.1,2
DNA-based technologies are complex and costly to perform and exhibit excellent sensitivity only with fresh or frozen tissue samples. However, such samples are often blindly obtained with no prior knowledge of the type, quantity, and viability of tumor cell nuclei. A more pragmatic approach to tissue selection and biomarker expression is required for developing selective and specific molecularly targeted drugs. The use of formalin-fixed paraffin-embedded (FFPE) tissue section remains an attractive and practical sample source for both genetic and proteomic analysis, allowing preservation of tissue morphology and parallel assessment of tumor type and viability.3 DNA and RNA have been successfully extracted from FFPE tissue archived for several years in storage. However, formalin fixation and paraffin embedding frequently lead to DNA and/or RNA fragmentation and degradation during storage, affecting the quality of molecular tests. Comparative NGS studies have demonstrated a decrease in library yield and an increase in false positive single nucleotide variants from clinical FFPE samples.4 Thus, it is desirable to develop surrogate proteomic tests that will reflect genetic mutations, in lieu of NGS. Such tests should be verified to correlate with molecular methods and can supplant them for quicker turnaround time, lower expense, and wider availability. This new approach may significantly impact clinical drug development for all cancer types, particularly pediatric cancer, due to the scant tissue samples and the small market. In this perspective, we focus on traditional as well as more innovative protein expression methods that can be applied to routinely processed FFPE tissue sections.
Tissue microarray (TMA)
High-throughput array-based screening techniques performed on multi-tumor tissue or TMA blocks have revolutionized our approach to diagnostics, prognostics, and drug discovery in oncology. TMA may be produced from frozen tissue, paraffin-embedded cell lines, or FFPE tissue and can be stored for later use.5 TMAs are created by taking small tissue cores from parent FFPE blocks and constructing a new multi-tissue block. This can be done by implanting the cores into a new recipient paraffin block or using steel mold blocks and grids to create a new composite TMA block. The tissue cores range in size from 0.6 up to several millimeters in diameter and can be punched more than once from each donor block.6 TMA blocks are then processed to yield numerous evaluable specimen samples on a single glass slide, thus reducing experimentation costs. All types of in situ experiments at the DNA, RNA, or protein level can be performed simultaneously on the TMA slide, thereby increasing quality assurance and minimizing inter-run variability.7,8 The success of TMA experiments depends highly on proper parent tissue selection, proper core sizing and alignment, and careful mapping of each specimen’s identity and localization. Inadequate or improper core selection may potentially lead to loss of data due to tissue or tumor heterogeneity or heterogeneity of analyte expression.8 However, using double, triplicate, or multiple cores for each tumor specimen will increase test concordance to full tissue sections. When storing TMA blocks, it is essential to consider the age of the original parent tissue to prevent prolonged storage and loss of tissue preservation. Proper core size and spacing must be carefully selected, as small tissue core sizes and crowded cores may lead to loss of tissue during processing.5 Nevertheless, TMA tissue sections have been successfully applied to the high-throughput screening of molecular targets through multiplex immunohistochemistry (IHC), FISH, RNAscope, and protein profiling.
FFPE methods of protein expression
IHC
Standard IHC techniques can be applied to single-tissue or TMA tissue sections using validated antibodies directed against specific protein targets of interest on manual or automated staining platforms. These platforms provide the advantage of retaining tissue morphology. Automated staining instruments are routinely used in developing countries to ensure the quality and consistency of IHC staining. These automated systems allow for the high-resolution capture and cataloging of individual tissue images, which can be stored and remotely accessed from a centralized database. Patterns of immunoreactivity will be antibody and target-specific and can be graded based on the intensity and distribution of staining to reflect a semiquantitative method of target expression. In addition to its use as a diagnostic tool in oncopathology, IHC protein expression can also provide prognostic information and help select patients for targeted therapy. However, assessing protein expression by IHC has been plagued by problems of sensitivity, specificity, and staining variability. Formalin fixation may mask antigen epitopes of interest, and tissue viability, decalcification, and processing may also adversely affect epitope expression. Automated IHC instruments have incorporated various signal amplification methods to increase sensitivity, which are best appreciated in multiplex labeling (discussed below). Stringent validation and standardization of procedures will increase the reliability of IHC, offering an easier and more widely available method of protein detection.9
Molecular IHC
In precision medicine, IHC can detect upregulated proteins, such as Human Epidermal Growth Factor Receptor 2 (HER2), Epidermal Growth Factor Receptor (EGFR), and Platelet-Derived Growth Factor Receptor (PDGFR), as well as upregulated signaling pathway members such as serine/threonine-protein kinase B-Raf and overexpressed proteins derived from mutated or altered genes. Identifying such proteins can serve as surrogate tests instead of traditional DNA-based tests. For example, Neuroblastoma RAS Viral Oncogene Homolog (NRAS) protein expression has been shown to be highly sensitive and specific in detecting NRAS mutations in melanoma.10–12 Molecular-specific IHC has the advantage of direct visualization of tumor heterogeneity and the ability to detect the expressed protein in a very limited sample with a shorter turaround time. Immunohistochemical methods have been developed to detect protein products of gene fusions, that can be performed as surrogates to FISH methods. Expression of these proteins in diagnostic clinical specimens can also serve as prognostic or therapeutic indicators. For instance, the immunohistochemical expression of PD-L1 in tumor cells or tumor-infiltrating lymphocytes is commonly used, regardless of tumor type or location, as an eligibility indicator for immune checkpoint inhibitors.13 Novel IHC tests are being incorporated into daily clinical practice, particularly in managing breast, colon, and brain cancers. Adequate standardization of IHC testing protocols is important to maintain the conformity of the test and minimize interlaboratory variation, as results may vary widely depending on the choice of fixative, antibody manufacturer, and type of immunostaining methods.14
Multiplex IHC/immunofluorescence
Immunofluorescence (IF) is a related immunological technique that can also be performed on FFPE tissue. IF allows for better signal detection with fewer steps, offering greater sensitivity than IHC.15 The development of multiplex IHC or Immunofluorescence (IF) for dual or multiple color coding has the potential to enhance the diagnostic yield and specificity of biomarker detection. Revolutionary multi-analyte IHC interrogates multiple targets on a single slide, maximizing the use of scarce tissue specimens. The methodology involves simultaneous or sequential staining steps and often employs signal amplification aided by powerful automated multispectral imaging software (Fig. 1).16 In IF, fluorescent detection can substitute for chromogen detection, facilitating easy multiplexing with fewer steps, better target colocalization, and a higher dynamic range.17 Hapten labeling, Opal multiplex, and Ventana Discovery-Ultra are commercially available methods that offer various advantages in multiplex IHC. These techniques are complemented by powerful image analysis that provides spatial localization and target quantification. Sequential rounds of staining, signal removal, and restaining have been employed with conventional fluorescence techniques in iterative indirect IF imaging to detect up to a 40-plex protein readout. Neogenomics MultiOmyx, using dye inactivation chemistry, can evaluate the co-expression of up to 60 biomarkers on a single slide.18,19
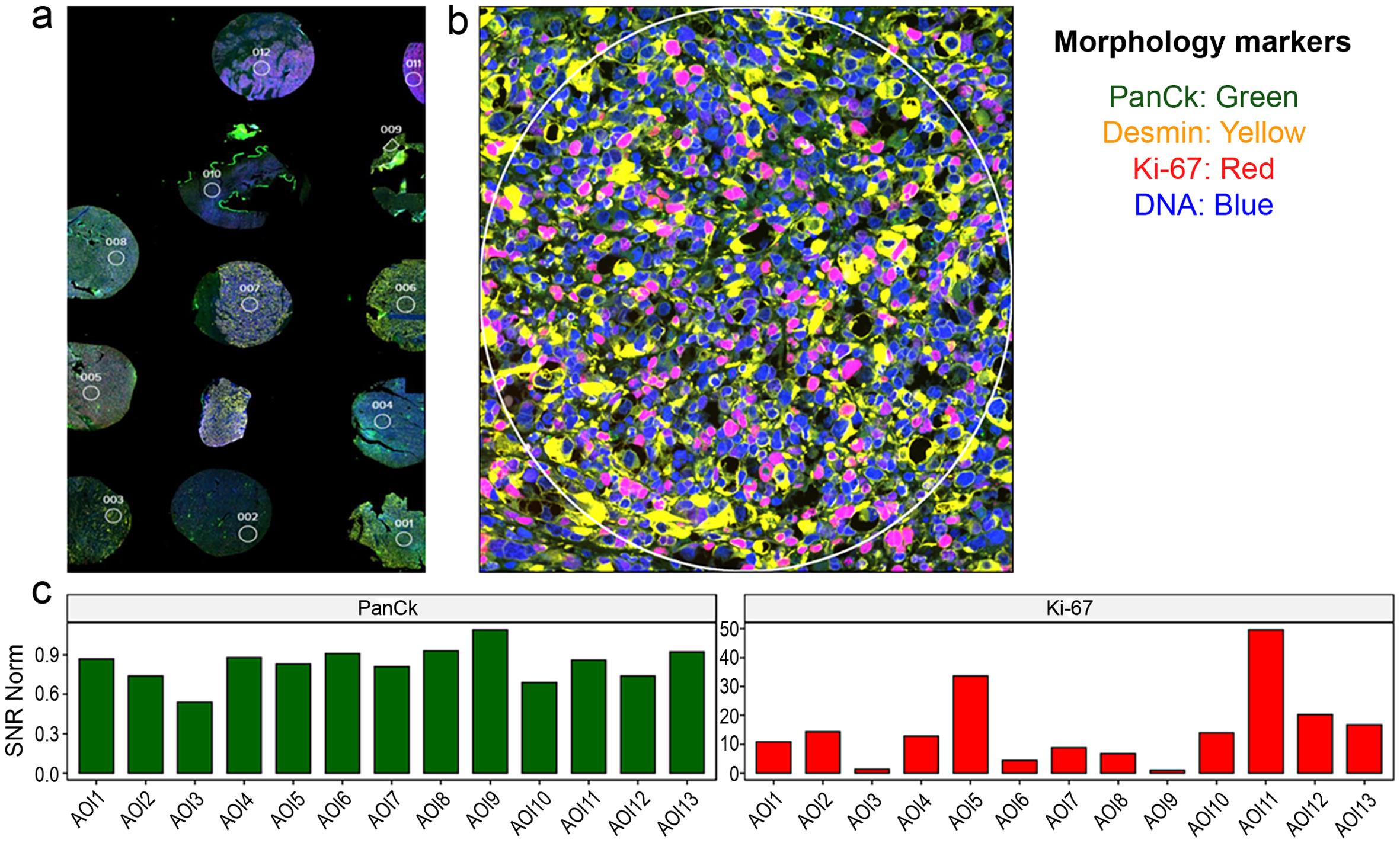
(a) Tissue microarray (TMA) slide of rhabdomyosarcoma stained with four different markers using immunofluorescence (mag ×40). (b) One selected core from the TMA in higher magnification shows colocalization of the stained markers. Tumor cells exhibit cytoplasmic staining with desmin and nuclear staining with Ki-67 and DNA marker. PanCK staining was not detected. (c) Imaging software can provide quantitative or semiquantitative data on the degree of expression of a specific protein target based on the color signal. In the TMA, Ki-67 staining was high in cases 5 and 11 (color signal quantified as 30 and 50 respectively) and lowest in cases 3 and 9 (color signal 0-1). In contrast, PanCK staining was nonexistent or low (color signal 0-0.9). Signal-to-Noise Ratio (SNR) indicates the color signal on the Y-axis.
Mass spectrometry
Mass spectrometry can be applied to tissue sections for the simultaneous detection of protein biomarkers in a hybrid histologic or IHC method combined with mass spectrometry (MS). High-throughput MS enables the extraction of molecular profiles from specific regions of FFPE tissue.20,21 When combined with IHC, multiple antibodies are tagged with metal isotopes of known molecular mass and detected with imaging mass spectrometry (IMS). IMS can localize panels of biomolecules in tissues and visualize the spatial distribution of biomarkers, proteins, and metabolites by their molecular masses. Various modalities of this laborious method require specialized instrumentation but can yield information on numerous tissue targets.22,23 MS can also be combined with laser capture microdissection to isolate or capture specific cells of interest in a complex tissue section under microscopic visualization. Matrix-assisted laser desorption/ionization combined with IMS is a more sensitive and robust technique for the in situ analysis of proteins from FFPE tissue sections, offering protein expression information on hundreds of analytes.23 MS-based technologies promise a significant impact on clinical, pharmacological, and tissue toxicodynamic research by providing molecular information from specific cell types within tissue sections.20
Digital spatial profiling (DSP)
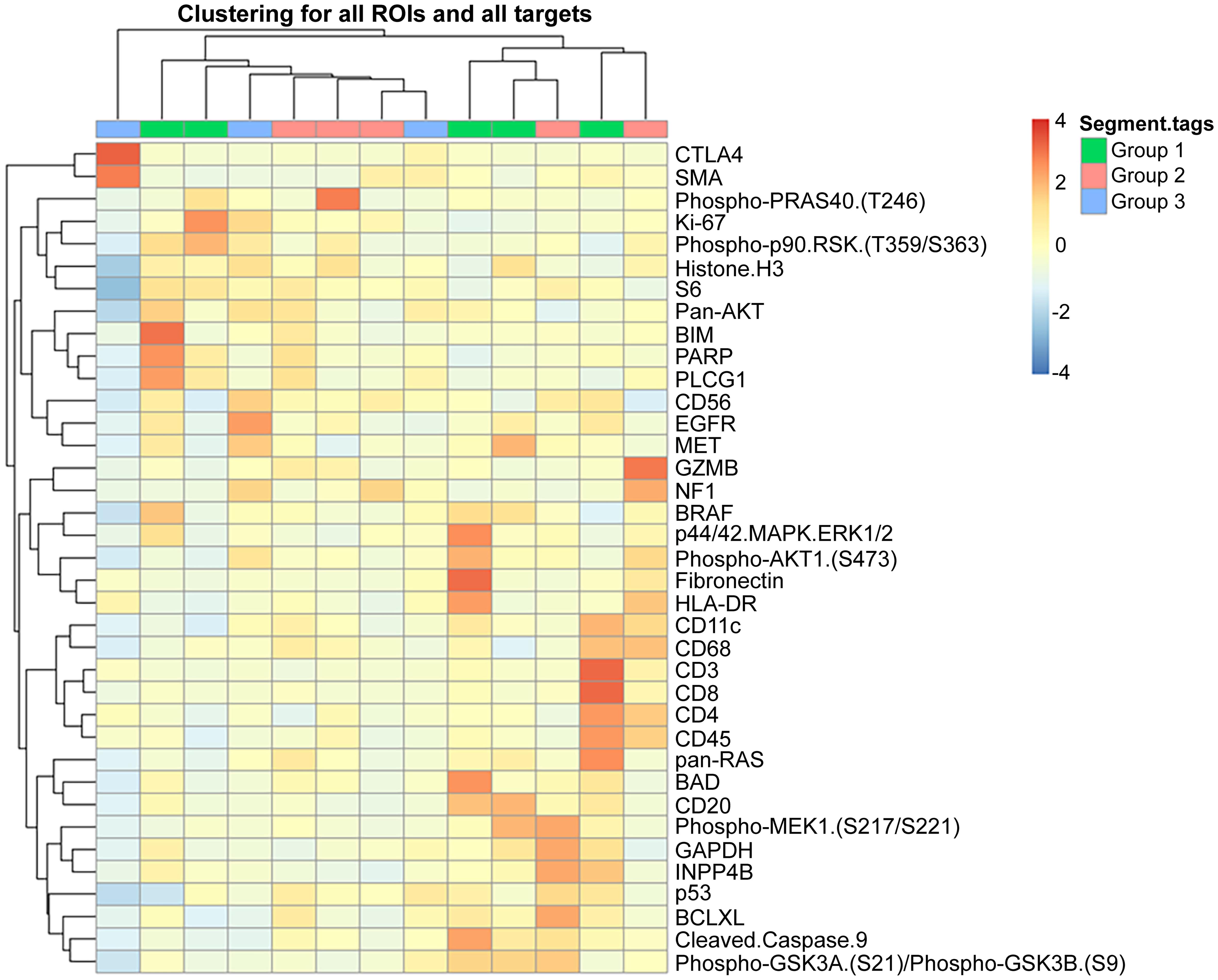
DSP is an integrated system for high-plex spatial profiling of proteins from FFPE tumor sections, offering the ability to analyze hundreds of protein targets. This platform, recently developed by NanoString Inc (www.nansotring.com ), employs digital barcoding technology and analytic software for spatial mapping of proteins. In this technique, target antibodies are covalently linked to DNA-indexing oligonucleotides with a UV-cleavable linker and applied to a specific area of interest. UV light liberates the indexing oligonucleotides, which are then collected and digitally counted.24 Prior immunolabeling of targets to identify regions of interest in tumor sections can help in the spatial mapping of proteins, reflecting intra-tumoral heterogeneity and tumor-microenvironment interactions. DSP has been particularly useful in immunotherapy to provide information on tumor-immune cell interactions (Fig. 2).25 It enables the identification and analysis of the whole transcriptome in a specific tumor region, tumor periphery, or stromal cells interacting with tumors.26 A recent modification of the technology allows morphologic-based spatial resolution of RNA and proteins in FFPE tissue at the subcellular level with high sensitivity. This advancement has the potential to generate three-dimensional localization of analytes, offering a more precise analysis of microenvironment and tumor interaction.27
Clinical applications
Apart from the traditional single antibody-single slide IHC, most of the new advances in tissue section proteomics are not widely available and remain at the research level. Few laboratories have incorporated multiplex IHC/IF in routine clinical laboratories, while the use of MS hybrid techniques is still hampered by cost and methodological challenges (Table 1). DSP is a promising alternative technique with the potential to advance rapidly in the clinical field. DSP has been successfully used to detect fusion transcripts in clinical samples of carcinomas and sarcomas.28 While the cost of whole transcriptome analysis ranges in the thousands, the DSP method can be modified to detect a specific panel, thereby reducing the cost and turnaround time to a few days. As an innovative example of tissue section proteomics, DSP methods can provide abundant information from FFPE tissue and detect analytes that may later be incorporated into routine use by IHC. Both DSP and multiplex molecular IHC/IF offer vast clinical potential and can play a significant role in precision medicine and therapy.
Comparison of different protein detection methods used on FFPE
| Single plex IHC | Multiplex IHC | Digital spatial profiling | Mass spectrometry | |
|---|---|---|---|---|
| Principle | Immunologic antigen-antibody reaction | Sequential immunologic reactions, computerized data display | Hybrid immunologic and probe hybridization reactions | Hybrid histologic, immunologic and mass spectrometry |
| Technique Complexity | Simple | Complex, needs specialized training | Complex, needs specialized training | Laborious based on preparation methods |
| Availability & Clinical Applications | Widely available with more test potential | Becoming more widely available; offered commercially; Emerging clinical use | Restricted to specialized centers for research; offered commercially; Vast research and clinical potential | Restricted to specialized centers for research; Requires method improvement |
| Interpretation | Subjective; preserves tissue morphology | Semiautomated; preserves tissue morphology | Fully automated; preserves tissue morphology | Fully automated; Can target specific area through microdissection |
| Cost | + (affordable) | ++ | +++ | +++ (expensive) |
Cancer precision proteomics, which focuses on identifying molecularly targeted proteins and signaling pathways involved in tumorigenesis, has altered the approach to clinical cancer drug development from DNA to protein testing. In hospital pathology laboratories, most tumor tissue is processed for clinical use and is only available as FFPE tissue. Novel technologies have been developed to identify protein targets in FFPE cancer tissue sections, providing more insights into drug discovery. In clinical settings, therapeutic targets in common epithelial and mesenchymal cancers are currently detected through nucleic acid-based sequencing. However, due to the poor quality of DNA extracted from regular FFPE tissue blocks, alternative approaches to formalin fixation and paraffin embedding have been developed to reduce the impact of artifacts. Acid-deprived formalin, gelatin embedding, and undecalcified bone embedding hold promise for more preserved extracted DNA, better performance, and more optimal nucleic acid-based research.29,30 However, more research is needed to evaluate the impact of these novel FFPE modifications on IHC and other protein expression techniques. Tissue section proteomics and FFPE tumor sampling have ushered in a new era of morphologic and spatial biology, offering a practical solution for the development of molecularly targeted drugs in cancer.
Declarations
Acknowledgement
None.
Funding
None.
Conflict of interest
The authors declare no conflict of interest.
Authors’ contributions
Conceptualization: AA, AO; Data curation: AO; Formal Analysis: AO, AA; Funding acquisition: Not applicable; Investigation: AA, AO; Methodology: AA, AO; Project administration: AA; Resources: AA; Software: AA; Supervision: AA; Validation: AA, AO; Visualization: AA, AO; Writing – original draft: AA; Writing – review & editing: AA, AO.

 Author information
Author information