Introduction
Pharmaceutical excipients are ideally anticipated to be pharmacodynamically inert, and they are used to formulate, stabilize, deliver, and preserve the active pharmaceutical ingredient (API). However, like the API, excipients may also have their own bioactivity.1–4 Many excipients used in pharmaceutical formulations are generally recognized as safe (GRAS) compounds in the food industry, and trust in these compounds may be conferred by their long history of use in consumer products and various pharmaceutical formulations.5–9 In both Europe and the United States, formulations with commercially established excipients may be approved without the need for the full, excipient-specific toxicology report that is required for new chemical entities. Similarly, excipients with prior use in Japan are not evaluated at the same level as novel excipients without prior use.10
Trehalose is a disaccharide that is used in oral and intravenous pharmaceutical formulations. It is a nonreducing disaccharide formed from two α-glucose units joined by a 1-1 glycosidic bond. The alpha-1,1- isomer is quite common across fungi, plants, and insects, where it is suggested that trehalose is used as a natural energy storehouse or cryptobiotic.11,12 Trehalose synthesis is absent in vertebrates, and there are no trehalose synthesis genes in vertebrates.13 However, the trehalose-catabolizing enzyme trehalase (EC 3.2.1.28) is reported in vertebrates, including mammals, where it is anchored to the brush border membrane of the intestine and kidney epithelium.14 The nonreducing alpha-1,1-glycosidic bond makes trehalose an exceptionally stable sugar that can withstand acid hydrolysis and cleavage by alpha-glycosidases. The crystalline dihydrate trehalose does not readily absorb water and is stable up to 97°C. Compared to sucrose (another disaccharide with a similar molecular weight), trehalose has lower solubility, a lower diffusion coefficient, a higher hydration number, and a higher viscosity. Trehalose can stabilize proteins or lipid preparations through direct interactions by hydrogen bonding to the hydrophilic protein surface or indirectly by binding water.6–9,15
The desirable physical properties of trehalose make it an ideal excipient in drug formulations; hence, it is used in approved medicinal products, such as vaccines for ovine rinderpest and mammalian cell culture desiccation.16,17 Recently, trehalose has been used as an excipient for generating monoclonal antibodies (Genetech’s Herceptin, Avastin, and Lucentis) and recombinant protein (Baxter’s Advate).15 Given that trehalose’s GRAS status is based on its historical use as a food additive (GRAS Notice No. 912), it is important to study the bioactivity of trehalose, especially when it bypasses the intestine through intravenous and intramuscular injections. When trehalose is used in oral formulations, it is hydrolyzed by trehalase, and the potential impact of this metabolism on gastrointestinal physiology needs critical assessment.
Rodent models, especially mice, are the most common animals used in biomedical research studies. While the use of rodent models has been indispensable, we have found that these models do not always translate well to humans.18 This may be due to physiological differences.19,20 Hence, in this study, using in silico methods, we screened the protein targets of trehalose in Homo sapiens, Mus musculus, and Rattus norvegicus (hereafter human, mouse, and rat) for a comparative and evidence-based assessment of its network pharmacology and pharmacodynamic properties.
Materials and methods
The SMILES structure of trehalose acquired from the PubChem database was input into the SwissTargetPrediction database to identify human-, mouse-, and rat-specific targets.21–25 The target list of trehalose was processed based on their probability scores to identify the highest affinity target(s), and only the targets with a probability score greater than 0 were used for further analysis (File S1). The enzyme trehalase (EC 3.2.1.28) was identified as a binding target by SwissTargetPrediction, but it has a probability of binding of 0, likely influenced solely by its enzymatic activity; hence, it was excluded as a binding target. The SwissADME database was used to assess the physiochemical and pharmacokinetic parameters of trehalose (Supplement S1).21 The targets of trehalose were subclassified into functional categories, and the relative proportion of each of the functional categories among the total number of targets was estimated and compared between human, mouse, and rat. This study did not involve the use of animals or the participation of human subjects and hence was not subjected to review by the relevant ethics committee.
We used the R package HPAnalyze to assess the tissue and subcellular expression (protein and mRNA) of trehalose targets in humans from the Human Protein Atlas (File S1).22,26,27 The data for protein, mRNA and subcellular expression were graphed using ggplot2.28 Protein expression was profiled under the following four categories: high, medium, low and no expression. The mRNA expression was categorized based on protein transcripts per kilobase million (pTPM) ranging from 0 to 1,250. While the subcellular locations of the trehalose targets were dichotomized as absent or present. The expression of trehalase protein in human tissues was also assessed and categorized as high, medium, or low (File S1). The affinity of trehalose for human-, mouse-, and rat-specific trehalases was assessed by molecular docking using AutoDock vina 1.2.0 as reported previously.25,29,30 The predictive toxicity assessment of trehalose was performed using ProTox-II tox-new.charite.de (File S1).31
Results
Trehalose is used in oral, intravenous, and intramuscular formulations due to its wider and compatible pharmacokinetic properties (Table 1). The low lipophilicity (iLOGP = 0.57, consensus Log P = −3.55) and high water solubility (ESOL Log S = 0.94) of trehalose support its use in a diverse range of formulation types. The pharmacokinetic parameters suggest that trehalose cannot easily cross the plasma membrane unless facilitated by a specific or selective transporter. Predictably, trehalose has a low bioavailability (0.17) and low gastrointestinal absorption, and it is impermeable to both the blood-brain barrier and skin barrier (log Kp = −11.36 cm/s). Trehalose is also an observed substrate for p-glycoprotein, which further decreases its gastrointestinal absorption. Trehalose did not appear to inhibit cytochrome liver enzymes (CYP1A2, CYP2C19, CYP2C9, CYP2D6, or CYP3A4) assessed in this study, suggesting that trehalose is a favorable excipient that will not affect the pharmacokinetics of the API in the formulation. Additionally, we assessed the predictive toxicity of trehalose and failed to observe any concerning toxicity features, further validating the favorable feature of trehalose as an excipient (Supplementary file). The predictive toxicity assessment showed an LD50 of 29.7 g/kg and a toxicity class of 6 with a prediction accuracy of 72.9%.
Physicochemical and pharmacokinetic properties of trehalose
| Property | Trehalose |
|---|---|
| Physicochemical Properties | |
| MW | 342.3 |
| #Heavy atoms | 23 |
| #Aromatic heavy atoms | 0 |
| Fraction Csp3 | 1 |
| #Rotatable bonds | 4 |
| #H-bond acceptors | 11 |
| #H-bond donors | 8 |
| Lipophilicity | |
| iLogP | 0.57 |
| Consensus Log P | −3.55 |
| Water Solubility | |
| ESOL Log S | 0.94 |
| Ali Class | Highly soluble |
| Pharmacokinetics | |
| GI absorption | Low |
| BBB permeant | No |
| Pgp substrate | Yes |
| CYP1A2 inhibitor | No |
| CYP2C19 inhibitor | No |
| CYP2C9 inhibitor | No |
| CYP2D6 inhibitor | No |
| CYP3A4 inhibitor | No |
| Log Kp (cm/s) | −11.36 |
| Druglikeness | |
| Bioavailability Score | 0.17 |
Trehalose is metabolized by trehalase in mammals; therefore, tissue-specific expression of this enzyme in human was assessed. Trehalase was moderately expressed in the kidney, duodenum, and small intestine, while its expression in the ovary, adrenal gland, colon and cerebral cortex was low (Table 2). This finding suggested that trehalose is preferentially metabolized in the kidneys, duodenum, and small intestine; therefore, trehalose may not be a preferred excipient if the intention is to deliver the API to these target organs. We also observed considerable variability in the affinity of trehalose to human, mouse, and rat trehalases. Mouse trehalase had the highest affinity for trehalase, followed by human trehalase, and least of all rat trehalase (Table 2). This significant species-specific variability in enzyme-substrate affinity has become important in drug discovery studies. Hence, considerable caution is necessary when extrapolating findings from preclinical rodent studies to design phase 1 clinical trials involving human subjects.
Protein expression levels of trehalase in human and the affinity* of trehalose to human-, rat- and mouse-specific trehalases
| Tissue | Cell type | Level |
|---|---|---|
| Adrenal gland | Glandular cells | Low |
| Cerebral cortex | Neuronal cells | Low |
| Colon | Peripheral nerve/ganglion | Low |
| Duodenum | Glandular cells | Medium |
| Kidney | Cells in tubules | Medium |
| Ovary | Follicle cells | Low |
| Small intestine | Glandular cells | Medium |
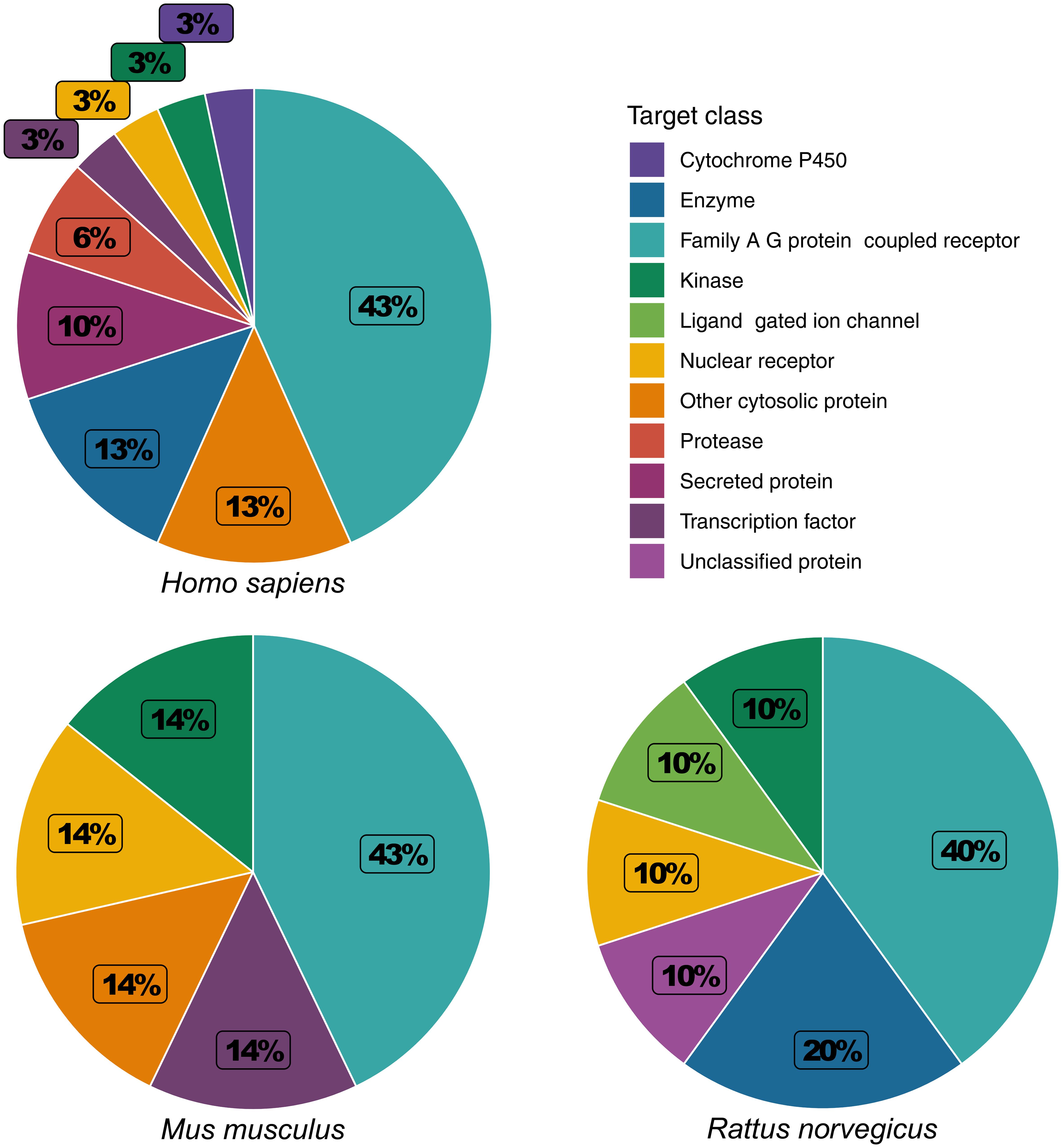
The species-specific variability of trehalose was further evident from differences in its binding targets in human, mouse, and rat. Family A G protein-coupled receptors (GPCRs) made up the largest portion of the binding target classes of trehalose. This was conserved across human (43%), mouse (43%), and rat (40%) (Fig. 1). However, the nature of the other target classes of trehalose differed considerably among the three species. In human, the next three most prominent target classes were enzymes (13%), other cytosolic proteins (13%), and secreted proteins (10%). Other cytosolic proteins were also identified as a prominent target class in mouse (14%), and enzymes were identified as a prominent target class in rat (20%). Secreted proteins were not identified as targets in either mouse or rat. In mouse, the remaining target classes were kinases (14%), nuclear receptors (14%), and transcription factors (14%). This was not conserved between human and mouse. Kinases, nuclear receptors, and transcription factors each made up 3% of the binding targets in human. In rat, the remaining target classes were kinases (10%), ligand-gated ion channels (10%), nuclear receptors (10%), or unclassified proteins (10%). Unclassified proteins were not identified as binding targets in human. This data suggest that protein binding targets largely differ between humans and rodent models. Therefore, preclinical safety and efficacy assessments of trehalose using rodent models may not be relevant for extrapolating the findings to design human clinical trials.
Variations in the binding targets of trehalose were observed among human, mouse and rat (Table 3), once again highlighting the potential irrelevance of rodent models in assessing trehalose-containing formulations for therapeutic applications in human. Cyclin-dependent kinase 1 (CDK1) is a serine/threonine protein kinase involved in cell cycle regulation and transcription at the G1 checkpoint,32 and it had the strongest binding affinity for trehalose (probability score = 0.24). CDK1 and heat shock protein HSP 90-alpha (HSP90AA1) were predicted binding targets of trehalose in both human and mouse, but not in rat. Dopamine D2 receptor (DRD2) was a predicted binding target of trehalose in human, mouse, and rat with similar low affinities. Dopamine D1 receptor (DRD1) was observed in only human and mouse. Dopamine D3 receptor (DRD3) was observed in only human and rat. While all serotonin (5-HT) receptors were predicted targets of trehalose in human, mouse, and rat, different receptors and subunits are targeted. Serotonin 1 receptor was observed in both human (HTR1B) and rat (HTR1A and HTR1B), but not mouse. Serotonin 2 receptor was observed in human (HTR2A, HTR2B, and HTR2C), mouse (HTR2C), and rat (HTR2A). Serotonin 6 receptor (HTR6) was only observed in human. RAR-related orphan receptor gamma (RORC) and STAT3 were predicted binding targets of trehalose in human and mouse, but not in rat. Alpha adrenergic receptors were predicted binding targets of trehalose in human and rat, but not in mouse. The alpha 1a adrenergic receptor (ADRA1A) was observed in both human and rat. Alpha 1b adrenergic receptor (ADRA1B) was observed in only rat, and the alpha 1d adrenergic receptor (ADRA1D) was observed in only human. The alpha 2b and 2c adrenergic receptors (ADRA2B and ADRA2C) were observed in both human and rat. Glutamate carboxypeptidase II (FOLH1) was a predicted binding target of trehalose in human and rat, but not mouse.
The binding targets of trehalose in Homo sapiens, Mus musculus, and Rattus norvegicus
| Target protein | Gene | Target protein class | Probability |
|---|---|---|---|
| Homo sapiens | |||
| Cyclin-dependent kinase 1 | CDK1 | Kinase | 0.24 |
| Heat shock protein HSP 90-alpha | HSP90AA1 | Other cytosolic protein | 0.11 |
| Vascular endothelial growth factor A | VEGFA | Secreted protein | 0.11 |
| Gamma-secretase | PSEN2 | Protease | 0.11 |
| Gamma-secretase | PSENEN | Protease | 0.11 |
| Gamma-secretase | NCSTN | Protease | 0.11 |
| Gamma-secretase | APH1A | Protease | 0.11 |
| Gamma-secretase | PSEN1 | Protease | 0.11 |
| Gamma-secretase | APH1B | Protease | 0.11 |
| Acidic fibroblast growth factor | FGF1 | Secreted protein | 0.11 |
| Heparanase | HPSE | Enzyme | 0.11 |
| Basic fibroblast growth factor | FGF2 | Secreted protein | 0.10 |
| Galectin-4 | LGALS4 | Other cytosolic protein | 0.10 |
| Galectin-3 | LGALS3 | Other cytosolic protein | 0.10 |
| Galectin-8 | LGALS8 | Other cytosolic protein | 0.10 |
| Glutamate carboxypeptidase II | FOLH1 | Protease | 0.10 |
| Serotonin 2b (5-HT2b) receptor | HTR2B | GPCR (Family A) | 0.10 |
| Alpha-2a adrenergic receptor | ADRA2A | GPCR (Family A) | 0.10 |
| Adrenergic receptor alpha-2 | ADRA2C | GPCR (Family A) | 0.10 |
| Alpha-2b adrenergic receptor | ADRA2B | GPCR (Family A) | 0.10 |
| Dopamine D1 receptor | DRD1 | GPCR (Family A) | 0.10 |
| Dopamine D2 receptor | DRD2 | GPCR (Family A) | 0.10 |
| Alpha-1d adrenergic receptor | ADRA1D | GPCR (Family A) | 0.10 |
| Serotonin 2a (5-HT2a) receptor | HTR2A | GPCR (Family A) | 0.10 |
| Serotonin 2c (5-HT2c) receptor | HTR2C | GPCR (Family A) | 0.10 |
| Dopamine D3 receptor | DRD3 | GPCR (Family A) | 0.10 |
| Cytochrome P450 2D6 | CYP2D6 | Cytochrome P450 | 0.10 |
| Serotonin 6 (5-HT6) receptor | HTR6 | GPCR (Family A) | 0.10 |
| Alpha-1a adrenergic receptor* | ADRA1A | GPCR (Family A) | 0.10 |
| Serotonin 1b (5-HT1b) receptor* | HTR1B | GPCR (Family A) | 0.10 |
| Nuclear receptor ROR-gamma | RORC | Nuclear receptor | 0.10 |
| Signal transducer and activator of transcription 3 | STAT3 | Transcription factor | 0.10 |
| Sialidase 3 | NEU3 | Enzyme | 0.10 |
| Sialidase 2 | NEU2 | Enzyme | 0.10 |
| Sialidase 4 | NEU4 | Enzyme | 0.10 |
| Mus musculus | |||
| Cyclin-dependent kinase 1* | Cdk1 | Kinase | 0.24 |
| Heat shock protein HSP 90-alpha* | Hsp90aa1 | Other cytosolic protein | 0.11 |
| Dopamine D1 receptor* | Drd1 | GPCR (Family A) | 0.10 |
| Dopamine D2 receptor* | Drd2 | GPCR (Family A) | 0.10 |
| Serotonin 2c (5-HT2c) receptor* | Htr2c | GPCR (Family A) | 0.10 |
| Nuclear receptor ROR-gamma* | Rorc | Nuclear receptor | 0.10 |
| Signal transducer and activator of transcription 3* | Stat3 | Transcription factor | 0.10 |
| Rattus norvegicus | |||
| Glutamate carboxypeptidase II | Folh1 | Enzyme | 0.10 |
| Serotonin 1a (5-HT1a) receptor | Htr1a | GPCR (Family A) | 0.10 |
| Alpha-1b adrenergic receptor | Adra1b | GPCR (Family A) | 0.10 |
| Alpha-1a adrenergic receptor | Adra1a | Ligand-gated ion channel | 0.10 |
| Serotonin 1b (5-HT1b) receptor | Htr1b | GPCR (Family A) | 0.10 |
| Alpha-2c adrenergic receptor* | Adra2c | Nuclear receptor | 0.10 |
| Alpha-2b adrenergic receptor* | Adra2b | Unclassified protein | 0.10 |
| Dopamine D2 receptor* | Drd2 | GPCR (Family A) | 0.10 |
| Serotonin 2a (5-HT2a) receptor* | Htr2a | Kinase | 0.10 |
| Dopamine D3 receptor* | Drd3 | Enzyme | 0.10 |
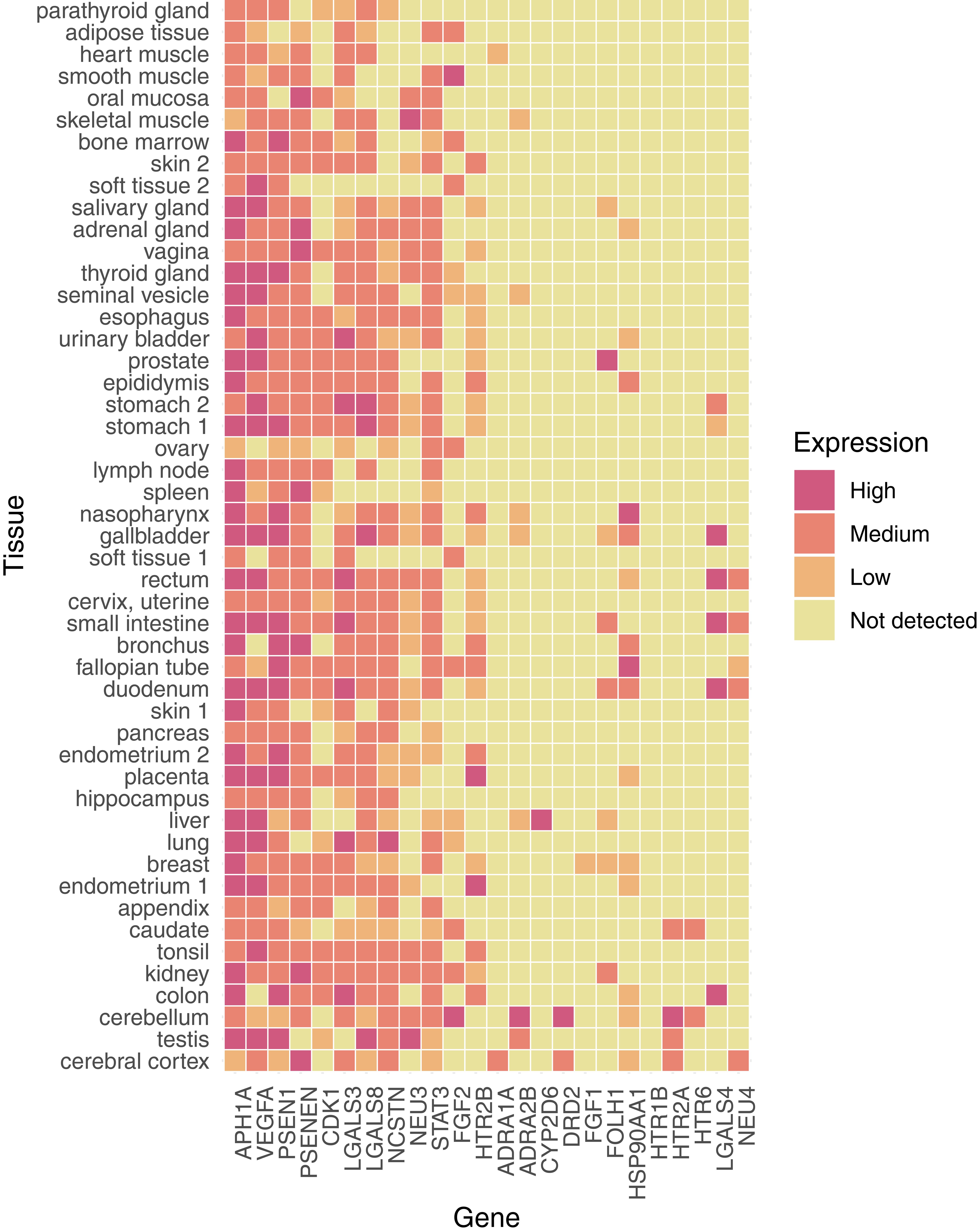
We assessed the protein expression profile of the targets of trehalose in human. The protein expression of trehalose targets was categorized into four categories – high, medium, low, and no expression (Fig. 2). Presenilin is a membrane-bound protease that makes up the catalytic unit of the gamma secretase complex, and together, these proteins may cleave amyloid precursor proteins into beta-amyloid chains.33 Generally, presenilin/gamma secretase-associated proteins had a high, widely distributed expression. The alpha-1 homolog A (APH1A) subunit had the highest expression across all body tissues, and it was at least moderately expressed in all tissues except for skeletal muscle, the ovary, and the cerebral cortex, where it had only low expression. Presenilin-1 (PSEN1) was widely expressed, but was absent from adipose and oral mucosa. In contrast, presenilin enhancer protein 2 (PSENEN) was absent from the parathyroid gland, lung, and testis. Nicastrin (NCSTN) was absent from adipose tissue, heart muscle, smooth muscle, oral mucosa, skeletal muscle, bone marrow, soft tissue, lymph nodes, or the spleen. At least three of the four observed presenilin/gamma secretase proteins were present in every tissue, except for adipose tissue and oral mucosa. There was also ubiquitously low expression of these presenilin/gamma secretase-associated proteins in the ovary. The binding affinity of trehalose to human, rat and mouse PSEN1 differed significantly and was observed to be 188.62, 142.74 and 114.74 µM, respectively.
The expression profile was categorized as follows: high (red), medium (orange), low (light orange) or no expression (yellow).
We observed three families of GPCR neurotransmitter receptors: the serotonin (5-HT) receptor family, the alpha adrenergic receptors, and the dopamine receptor family (Fig. 2). Serotonin 6 (HTR6) receptor had medium expression in the caudate nucleus and cerebellum. Serotonin 2a receptor (HTR2A) was also expressed in the cerebellum and caudate, as well as in the testis and cerebral cortex. Serotonin 2b receptor (HTR2B) was widely expressed throughout the skin, glands, gastrointestinal tract, and female reproductive tract. Alpha-1a adrenergic receptor (ADRA1A) was expressed only in heart muscle and the cerebral cortex. Alpha-2b adrenergic receptor (ADRA2A) was highly expressed in the cerebellum and moderately/lowly expressed in skeletal muscle, seminal gland, nasopharynx, gallbladder, liver and testis. Dopamine D2 receptor (DRD2) was highly expressed in the cerebellum and moderately expressed in the cerebral cortex.
Galectins are a family of alpha-galactoside carbohydrate binding proteins.34 Galectin-3 (LGALS3), galectin-4 (LGALS4), and galectin-8 (LGALS8) were observed (Fig. 2). Galectin-4 was highly expressed in a few select tissues (gallbladder, rectum, small intestine, duodenum, and colon). Galectin-3 and galectin-8 had similar expression patterns, but both were absent from soft tissue and the spleen. Between the two, they had either high or medium expression in the urinary bladder, stomach, gallbladder, rectum, small intestine, duodenum, lung, or colon—a pattern that overlaps with galectin-4. Notably, galactin-8 was highly expressed in the testis, but galectin-4 and galactin-3 were absent.
We observed salidase-3 (NEU3) and salidase-4 (NEU4), hydrolases that cleave the glycosidic bond in neuraminic acids (Fig. 2). Viral neuraminidase is a drug target for preventing the spread of influenza infection.35 Salidase-3 is highly expressed in skeletal muscle and the testis. Salidase-4 is moderately expressed in the rectum, small intestine, duodenum, and cerebral cortex and has low expression in the fallopian tube. Salidase-3 is absent from the cerebral cortex and hippocampus but has moderate expression in the cerebellum. On the other hand, salidase-4 has moderate expression in the cerebral cortex, and it is absent from the cerebellum and hippocampus.
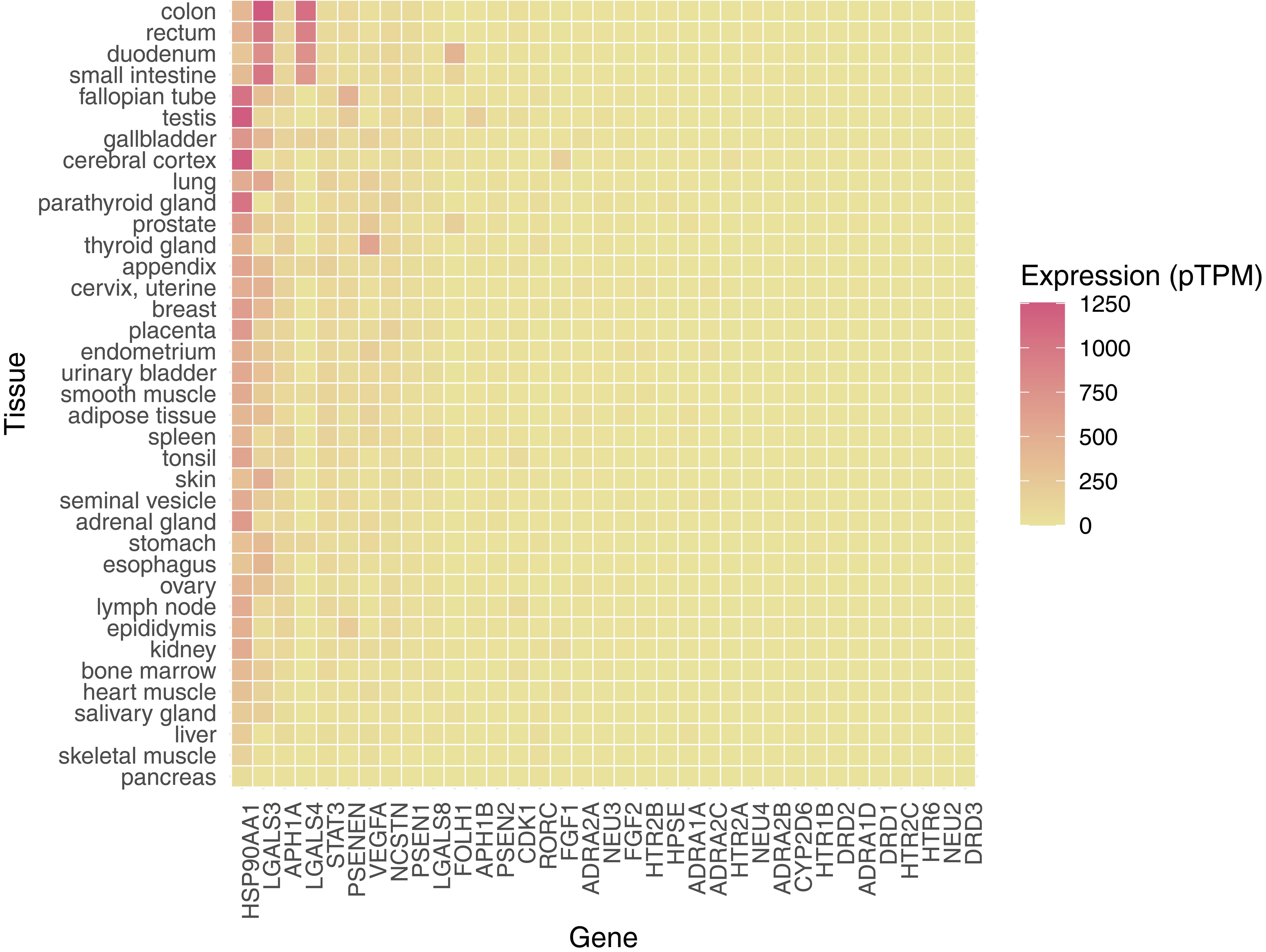
CDK1, which was the highest binding affinity target of trehalose in human and mouse (Table 3), has moderate expression across body tissues. Since CDK1 is involved in driving the cell cycle and cell proliferation,32 administration of trehalose-containing formulations may affect these tissues. The binding affinity of trehalose to human, rat and mouse CDK1 was observed to be 8.21, 8.21, and 10.66 µM, respectively. Vascular endothelial growth factor A (VEGFA) is a glycoprotein associated with neurons that promotes angiogenesis.36 VEGFA was widely expressed except for ovary, bronchus, and colon. It was highly expressed across reproductive organs, the GI tract, the liver, and the lung. It was only moderately expressed in the cerebral cortex, cerebellum, and hippocampus. The binding affinity of trehalose to human, rat and mouse VEGFA was very weak, and was observed to be 2,234.64, 2,697.50, and 2,697.50 µM, respectively. STAT3 is a transcription factor that is activated by Janus kinases in the JAK/STAT pathway.37 STAT3 was expressed at medium or low levels in the nervous system, muscle, glands, and gastrointestinal tract. Glutamate carboxypeptidase II (FOLH1) was expressed in the kidney, breast, liver, duodenum, small intestine, gallbladder, prostate, and salivary gland. HSP90AA1 was highly expressed in the fallopian tube and nasopharynx. Overall, the cerebral cortex, testis, and cerebellum tissues had the highest composite expression of binding targets of trehalose in human, followed by the target concentrations in the colon and kidney (Fig. 2). We were not able to obtain protein expression data for eleven proteins, namely, several alpha adrenergic receptors (ADRA1D, ADRA2A, and ADRA2C), two dopamine D receptors (DRD1 and DRD3), presenilin/gamma secretase (APH1B and PSEN2), a serotonin 2c receptor (HTR2C), sialidase 2 (NEU2), nuclear receptor ROR-gamma (RORC), and heparanase (HPSE). The RNA transcript expression of all the selected genes was graphed. Unsurprisingly, protein expression and RNA transcript expression data largely did not overlap with the available data (Figs. 2 and 3).
The expression profile was categorized based on the pTPM ranging from 0 to 1,250.
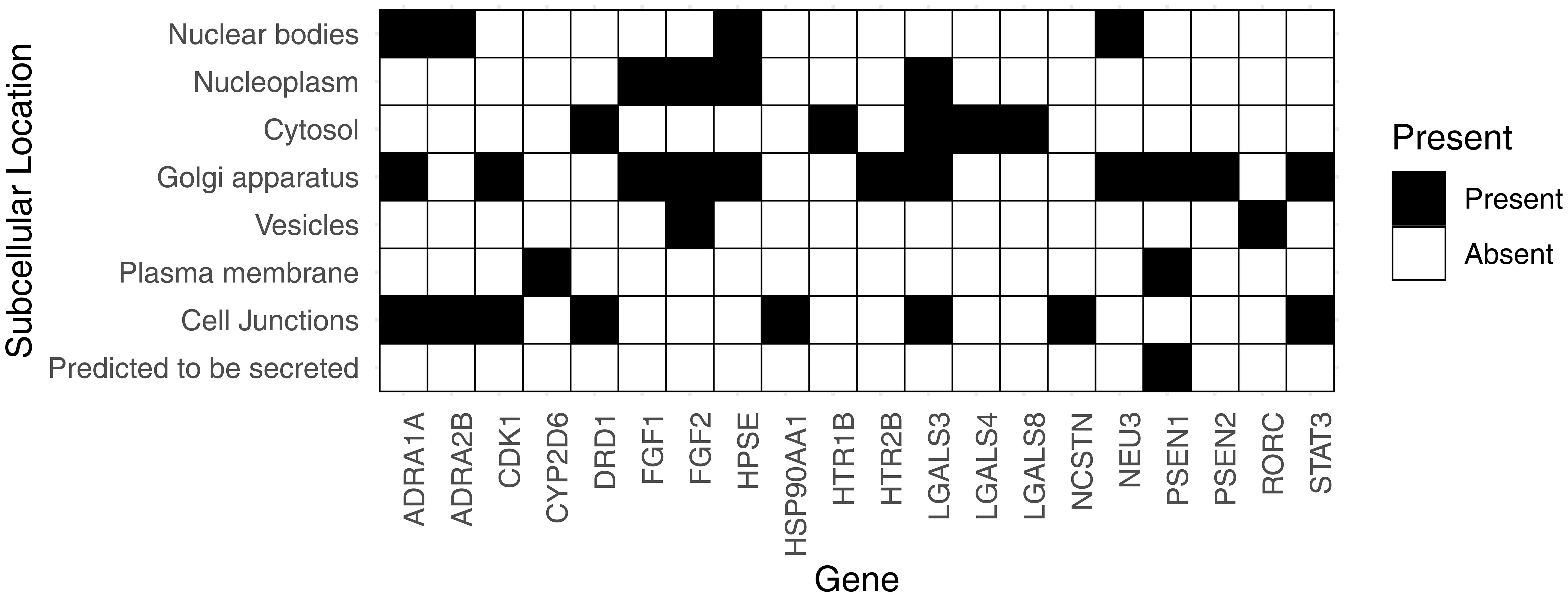
We also identified subcellular protein expression of the targets of trehalose in human. Since trehalose does not readily permeate the plasma membrane, we focused on proteins which are secreted or expressed on cell junctions, the plasma membrane, or secreted vesicles (Fig. 4). We observed that presenilin-1 was expressed on the plasma membrane and was also predicted to be secreted. Gamma-secretase, together with CDK1, is likely of considerable interest in assessing the safety and bioactivity of trehalose. Cytochrome P450 2D6 is the only other identified protein expressed on the plasma membrane; however, the affinity of trehalose for this target is very low. FGF3 and RORC were located in the vesicles. In addition, we observed a number of proteins expressed in the cell junction, including alpha-1a adrenergic receptor (ADRA1A), alpha-2b adrenergic receptor (ADRA2B), CDK1, dopamine D1 receptor (DRD1), heat shock protein HSP 90-alpha (HSP90AA1), galectin-3 (LGALS3), nicastrin (NCSTN), and STAT3. All these targets are highly relevant to the assessment of the bioactivity of trehalose in pharmaceutical formulations and hence should be integrated into drug development protocols.
Discussion
Trehalose is a GRAS compound which is widely used as an excipient in food products and pharmaceutical formulations due to its several desirable pharmacokinetic properties. The GRAS label for trehalose is largely based on historical evidence of its safety and a limited number of assessments using rodent and rabbit models.6–9,38–40 Although excipients are assumed to be pharmacologically inert, this can largely be attributed to a lack of suitable bioassays to quantify their pharmacodynamic effects in parallel to historical evidence of their safety. However, no chemical entity can be considered truly inert, and this especially applies to pharmaceutical excipients, which are often used in much higher concentrations than the API. Advancements in bioinformatics and cheminformatics tools address these gaps by making it feasible to objectively assess the pharmacodynamic activity of excipients with selected species specificity.41–45
This study highlights several pharmacodynamic features of trehalose which are relevant to its use in pharmaceutical formulations. The desirable physicochemical and pharmacokinetic properties of trehalose observed in this study are consistent with several previous in vivo studies,6,9,38,39,46,47 which validate its use as a pharmaceutical excipient. Among these desirable excipient properties are its lipophilicity, poor gastrointestinal permeability, slower metabolism, and negligible interference with liver cytochrome p450 enzymes. In addition, trehalose is also reported to enhance protein stability, which supports its preferential use in formulating biological therapeutics.6–9,48–50 The pharmacodynamic results are consistent with the in vivo findings, so this study validates the use of in silico tools in drug development.
Trehalose is selectively metabolized into monosaccharides by trehalase expressed in the kidney and intestine. The kinetics of this metabolic process are likely slow because the affinity of trehalose to human trehalase was observed to be approximately 66 µM. This observation is consistent with reports of peak trehalase concentration lasting for about 5 hours post intravenous administration in human, which is almost twice the duration typically reported for other disaccharides.8,39,51 We found significant inter-species variability in the affinity of trehalose to trehalase highlight the limitations of rodent studies as models for efficacy analysis of pharmaceutical formulations containing trehalose and perhaps other excipients. We observed major expression of trehalase in the kidneys and gastrointestinal system (duodenum and small intestines), which is consistent with previous findings.52 These observations are critical for screening all parenteral formulations containing trehalose, as such formulations are preferentially eliminated by renal excretion; therefore such formulations should be critically monitored for its impact on renal physiology.
We observed significant variation in the target classes in human, mouse, and rat. While Family A GPCRs were the most targeted protein class, the remaining target classes considerably varied. The GPCR repertoire varies considerably among mammals; for example, the canine GPCR genome is more closely related to that of human than to that of mouse and rat.53 Only 50% of GPCRs have a one-to-one ortholog between human and rat, as well as only 70% between mouse and rat, and the authors suggest that the GPCR repertoire may be more highly diverged than that of other subsets of the genome.54 This is reflected in the serotonin receptors, a family of GPCRs in which only one of five receptors is predicted binding targets of trehalose. This further emphasizes the irrelevance of rodent models in efficacy and safety analysis in preclinical drug discovery and development programs.
Among the many targets of trehalose identified in this study, CDK1 had the highest affinity for trehalose. However, the expression of CDK1 under physiological conditions was observed to be medium to low in human tissues, providing further assurance to the safety of trehalose use in pharmaceutical formulations. This assurance of the safety of trehalose is consistent with the findings of several other in vivo studies.6,8,9,39,40,51,55 The number of low-affinity targets of trehalose determined by analysis of the expression level and subcellular localization of these targets further confirmed that the impact of trehalose interaction with these targets on systemic physiology will be negligible. However, some of the targets of trehalose identified in this study may become relevant under specific pathological conditions (where the targets are upregulated or when their sensitivity is enhanced), and studies should be focused on the efficacy and safety assessments of trehalose formulations.
In summary, the comparative pharmacodynamic analysis of trehalose highlights the limitations of rodent models, specifically mouse and rat, in translating findings for the development of human therapeutics. The in silico analyses performed in this study may provide valuable additions to drug development programs as refinement tools for efficiency and cost optimization. It will be interesting to see if in silico comparative pharmacodynamics can be integrated into preclinical programs to make necessary corrections to improve translatability. Without such an approach, rodent model-based preclinical programs add limited insight into drug discovery and development at the expense of time and cost.
Supporting information
Supplementary material for this article is available at https://doi.org/10.61474/ncs.2023.00012 .
File S1
Compound ID, IUPAC name, Simplified Molecular Input Line Entry System (SMILES), Physicochemical Properties (from SwissADME database) of Trehalose are detailed. All the targets of Trehalose identified in the Swiss Target Prediction database and their expression (protein or mRNA) in various tissue or subcellular levels are summarised.
(XLSX)
Abbreviations
- ADRA1A:
alpha 1a adrenergic receptor
- ADRA1B:
alpha 1b adrenergic receptor
- ADRA1D:
alpha 1d adrenergic receptor
- APH1A:
alpha-1 homolog A
- API:
active pharmaceutical ingredient
- BBB:
blood brain barrier
- CDK1:
cyclin-dependent kinase 1
- CYP:
cytochrome P
- DRD1:
dopamine D1 receptor
- DRD2:
dopamine D2 receptor
- DRD3:
dopamine D3 receptor
- FGF:
fibroblast growth factor
- GI:
gastrointestinal
- GPCRs:
G protein coupled receptors
- GRAS:
generally recognized as Safe
- H:
hydrogen
- HSP:
heat shock protein
- JAK:
janus kinase
- MW:
molecular weight
- NCSTN:
nicastrin
- NEU3/4:
salidase-3/4
- PSEN1:
presenilin-1
- PSENEN:
presenilin enhancer protein
- Pgp:
p glycoprotein.
- RAR:
retinoic acid receptor
- RORC:
retinoic acid receptor-related orphan receptor gamma
- STAT:
signal transmission and activator of transcription
- VEGF:
vascular endothelial growth factor
- pTPM:
protein transcripts per kilobase million
- 5-HT:
serotonin
Declarations
Acknowledgement
None.
Funding
Research support from the University College Dublin-Seed Funding/Output-Based Research Support Scheme (R19862, 2019) and Stemcology (STGY2917, 2022) is acknowledged.
Conflict of interest
The authors declare that they have no conflicts of interest related to this publication.
Authors’ contributions
Conceptualization: AHSK; Data curation: JF, AHSK; Formal Analysis: JF, AHSK; Funding acquisition: AHSK; Investigation: JF, AHSK; Methodology: JF, AHSK; Project administration: AHSK; Resources: AHSK; Supervision: AHSK; Writing – original draft: JF, AHSK; Writing – review & editing: JF, AHSK.

 Author information
Author information